| functional annotation |
| Function |
Major facilitator superfamily protein |




|
| GO BP |
|
GO:0015904 [list] [network] tetracycline transmembrane transport
|
(2 genes)
|
ISS
|
 |
|
GO:0006855 [list] [network] xenobiotic transmembrane transport
|
(6 genes)
|
ISS
|
 |
|
GO:0098656 [list] [network] monoatomic anion transmembrane transport
|
(18 genes)
|
ISS
|
 |
|
GO:0010228 [list] [network] vegetative to reproductive phase transition of meristem
|
(287 genes)
|
IEA
|
 |
|
GO:0055085 [list] [network] transmembrane transport
|
(525 genes)
|
IEA
|
 |
|
GO:0006970 [list] [network] response to osmotic stress
|
(883 genes)
|
IEA
|
 |
|
GO:0009414 [list] [network] response to water deprivation
|
(1006 genes)
|
IEA
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(1086 genes)
|
IEA
|
 |
|
| GO CC |
|
GO:0005886 [list] [network] plasma membrane
|
(2529 genes)
|
ISM
|
 |
|
| GO MF |
|
GO:0008493 [list] [network] tetracycline transmembrane transporter activity
|
(3 genes)
|
ISS
|
 |
|
GO:0022857 [list] [network] transmembrane transporter activity
|
(1006 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001031360.1
NP_001323578.1
NP_001323579.1
NP_001323580.1
NP_001323581.1
NP_001323582.1
NP_001323583.1
NP_179291.2
|
| BLAST |
NP_001031360.1
NP_001323578.1
NP_001323579.1
NP_001323580.1
NP_001323581.1
NP_001323582.1
NP_001323583.1
NP_179291.2
|
| Orthologous |
[Ortholog page]
LOC103860499 (bra)
|
Subcellular
localization
wolf |
|
vacu 5,
plas 3,
E.R._vacu 3
|
(predict for NP_001031360.1)
|
|
vacu 5,
plas 3,
E.R._vacu 3
|
(predict for NP_001323578.1)
|
|
vacu 6,
plas 2,
mito 1,
extr 1,
E.R. 1
|
(predict for NP_001323579.1)
|
|
vacu 5,
plas 3,
E.R._vacu 3
|
(predict for NP_001323580.1)
|
|
plas 3,
vacu 3,
E.R. 2,
mito 1,
extr 1,
golg 1
|
(predict for NP_001323581.1)
|
|
plas 3,
vacu 3,
E.R. 2,
mito 1,
extr 1,
golg 1
|
(predict for NP_001323582.1)
|
|
vacu 5,
plas 3,
E.R. 1,
nucl_plas 1,
golg_plas 1,
cysk_plas 1,
mito_plas 1,
cyto_plas 1
|
(predict for NP_001323583.1)
|
|
vacu 5,
plas 3,
E.R._vacu 3
|
(predict for NP_179291.2)
|
|
Subcellular
localization
TargetP |
|
scret 9,
other 4
|
(predict for NP_001031360.1)
|
|
scret 9,
other 4
|
(predict for NP_001323578.1)
|
|
scret 9,
other 4
|
(predict for NP_001323579.1)
|
|
scret 9,
other 4
|
(predict for NP_001323580.1)
|
|
scret 9,
other 4
|
(predict for NP_001323581.1)
|
|
scret 9,
other 4
|
(predict for NP_001323582.1)
|
|
scret 9,
other 3
|
(predict for NP_001323583.1)
|
|
scret 9,
other 4
|
(predict for NP_179291.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with AT2G16990 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 5.1 |
AT1G74458 |
uncharacterized protein |
[detail] |
5007848 |
| 4.9 |
TMT2 |
tonoplast monosaccharide transporter2 |
[detail] |
829684 |
| 4.9 |
UMAMIT11 |
nodulin MtN21 /EamA-like transporter family protein |
[detail] |
818687 |
| 4.8 |
SWEET13 |
Nodulin MtN3 family protein |
[detail] |
835152 |
| 4.6 |
ZIFL2 |
zinc induced facilitator-like 2 |
[detail] |
823490 |
| 4.4 |
AT2G40460 |
Major facilitator superfamily protein |
[detail] |
818640 |
| 4.0 |
AT4G39795 |
hypothetical protein (DUF581) |
[detail] |
830138 |
|
Coexpressed
gene list |
[Coexpressed gene list for AT2G16990]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
263574_at
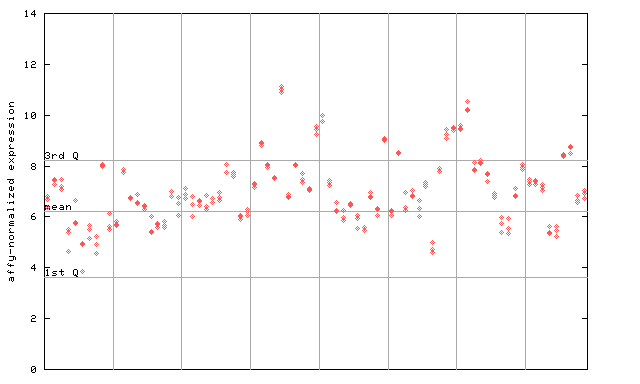
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
263574_at
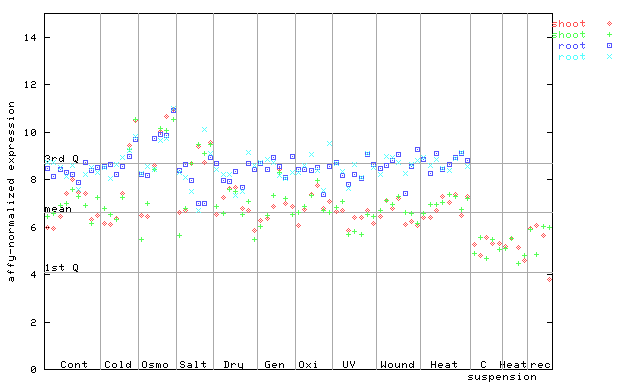
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
263574_at
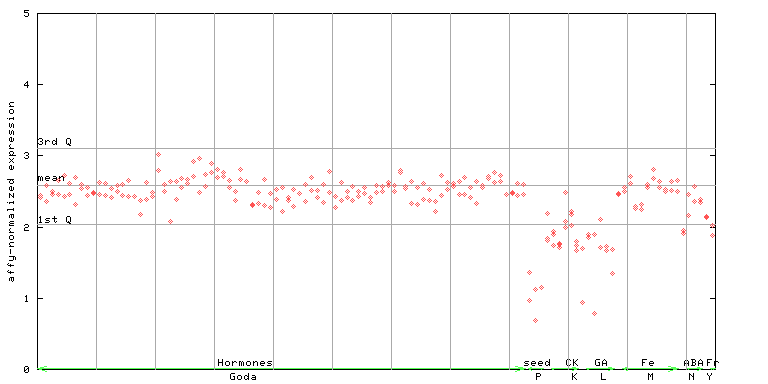
X axis is samples (xls file), and Y axis is log-expression.
|


















