| functional annotation |
| Function |
RING/U-box superfamily protein |




|
| GO BP |
|
GO:0016192 [list] [network] vesicle-mediated transport
|
(261 genes)
|
IEA
|
 |
|
GO:0044248 [list] [network] cellular catabolic process
|
(1086 genes)
|
IEA
|
 |
|
GO:1901701 [list] [network] cellular response to oxygen-containing compound
|
(1115 genes)
|
IEA
|
 |
|
GO:1901575 [list] [network] organic substance catabolic process
|
(1156 genes)
|
IEA
|
 |
|
GO:0098542 [list] [network] defense response to other organism
|
(2060 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001189695.1
NP_001318370.1
NP_001324231.1
NP_001324232.1
NP_001324233.1
NP_001324234.1
NP_001324235.1
NP_001324236.1
NP_001324237.1
NP_001324238.1
NP_001324239.1
NP_001324240.1
NP_001324241.1
NP_850278.1
NP_973618.1
|
| BLAST |
NP_001189695.1
NP_001318370.1
NP_001324231.1
NP_001324232.1
NP_001324233.1
NP_001324234.1
NP_001324235.1
NP_001324236.1
NP_001324237.1
NP_001324238.1
NP_001324239.1
NP_001324240.1
NP_001324241.1
NP_850278.1
NP_973618.1
|
| Orthologous |
[Ortholog page]
LOC547939 (gma)
LOC7457874 (ppo)
LOC7458651 (ppo)
LOC11425603 (mtr)
LOC11431193 (mtr)
LOC11432972 (mtr)
LOC100780416 (gma)
LOC100786013 (gma)
LOC100803627 (gma)
LOC101251216 (sly)
LOC103857659 (bra)
LOC103865579 (bra)
LOC112328435 (ppo)
|
Subcellular
localization
wolf |
|
nucl 8,
cyto_nucl 4,
extr 1,
vacu 1
|
(predict for NP_001189695.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001318370.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324231.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324232.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324233.1)
|
|
nucl 9,
chlo 1,
cysk 1
|
(predict for NP_001324234.1)
|
|
nucl 9,
chlo 1,
cysk 1
|
(predict for NP_001324235.1)
|
|
nucl 9,
chlo 1,
cysk 1
|
(predict for NP_001324236.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324237.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324238.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324239.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324240.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001324241.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_850278.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_973618.1)
|
|
Subcellular
localization
TargetP |
|
scret 7
|
(predict for NP_001189695.1)
|
|
other 8
|
(predict for NP_001318370.1)
|
|
other 8
|
(predict for NP_001324231.1)
|
|
other 8
|
(predict for NP_001324232.1)
|
|
other 8
|
(predict for NP_001324233.1)
|
|
other 5,
mito 4
|
(predict for NP_001324234.1)
|
|
other 5,
mito 4
|
(predict for NP_001324235.1)
|
|
other 5,
mito 4
|
(predict for NP_001324236.1)
|
|
other 8
|
(predict for NP_001324237.1)
|
|
other 8
|
(predict for NP_001324238.1)
|
|
other 8
|
(predict for NP_001324239.1)
|
|
other 8
|
(predict for NP_001324240.1)
|
|
other 8
|
(predict for NP_001324241.1)
|
|
other 8
|
(predict for NP_850278.1)
|
|
other 8
|
(predict for NP_973618.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with AT2G37150 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 7.1 |
ABA3 |
molybdenum cofactor sulfurase (LOS5) (ABA3) |
[detail] |
838224 |
| 6.3 |
AT4G32440 |
Plant Tudor-like RNA-binding protein |
[detail] |
829379 |
| 6.0 |
AT3G51130 |
uncharacterized protein |
[detail] |
824277 |
|
Coexpressed
gene list |
[Coexpressed gene list for AT2G37150]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
265470_at
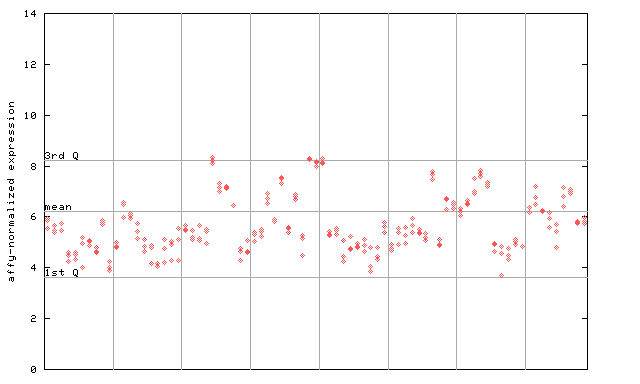
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
265470_at
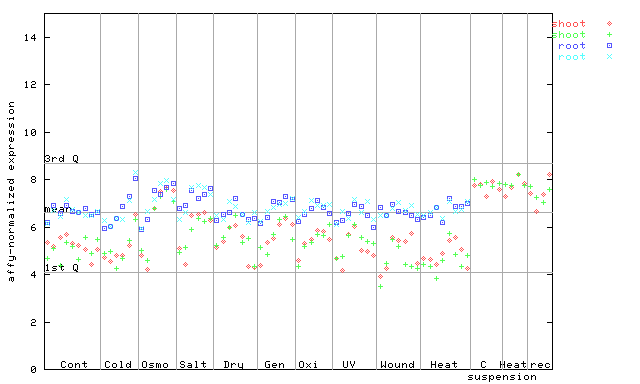
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
265470_at
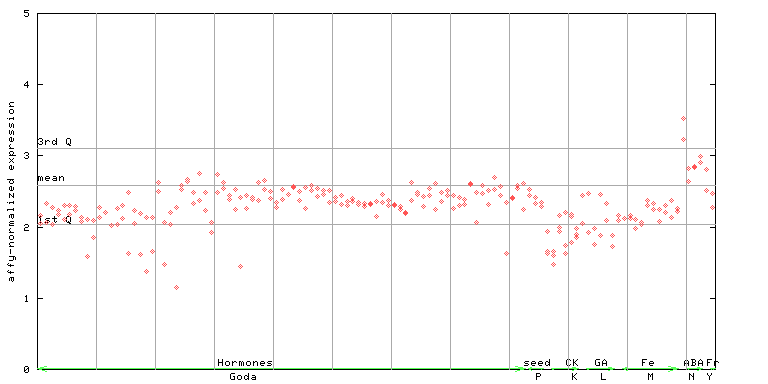
X axis is samples (xls file), and Y axis is log-expression.
|








