| functional annotation |
| Function |
N-MYC downregulated-like 2 |




|
| GO BP |
|
GO:2000012 [list] [network] regulation of auxin polar transport
|
(33 genes)
|
IMP
|
 |
|
GO:0009409 [list] [network] response to cold
|
(472 genes)
|
IDA
|
 |
|
GO:0009753 [list] [network] response to jasmonic acid
|
(611 genes)
|
IDA
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(1086 genes)
|
IDA
|
 |
|
| GO CC |
|
GO:0009507 [list] [network] chloroplast
|
(5004 genes)
|
ISM
|
 |
|
GO:0005737 [list] [network] cytoplasm
|
(13880 genes)
|
ISM
|
 |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001332289.1
NP_568251.1
|
| BLAST |
NP_001332289.1
NP_568251.1
|
| Orthologous |
[Ortholog page]
NDL1 (ath)
LOC4327306 (osa)
LOC4328772 (osa)
LOC4338041 (osa)
LOC4341312 (osa)
LOC7463744 (ppo)
LOC7470709 (ppo)
LOC11419160 (mtr)
LOC11426490 (mtr)
LOC18107680 (ppo)
LOC100781823 (gma)
LOC100782850 (gma)
LOC100783036 (gma)
LOC100793778 (gma)
LOC100804729 (gma)
LOC100815084 (gma)
LOC100817684 (gma)
LOC101255415 (sly)
LOC101262545 (sly)
LOC101266512 (sly)
LOC101290587 (tae)
LOC103845105 (bra)
LOC103849015 (bra)
LOC103850794 (bra)
LOC103855932 (bra)
LOC103856801 (bra)
LOC123060369 (tae)
LOC123068898 (tae)
LOC123144481 (tae)
LOC123160092 (tae)
LOC123165957 (tae)
LOC123404200 (hvu)
LOC123407859 (hvu)
LOC123442897 (hvu)
|
Subcellular
localization
wolf |
|
extr 4,
chlo 2,
vacu 2,
nucl 1
|
(predict for NP_001332289.1)
|
|
chlo 4,
nucl 1,
cyto 1,
plas 1,
cyto_nucl 1,
nucl_plas 1,
cyto_plas 1
|
(predict for NP_568251.1)
|
|
Subcellular
localization
TargetP |
|
scret 8,
other 4
|
(predict for NP_001332289.1)
|
|
other 8
|
(predict for NP_568251.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with NDL2 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 9.7 |
FLA15 |
FASCICLIN-like arabinogalactan protein 15 precursor |
[detail] |
824402 |
| 9.4 |
FLA16 |
FASCICLIN-like arabinogalactan protein 16 precursor |
[detail] |
818159 |
| 8.9 |
AT3G09540 |
Pectin lyase-like superfamily protein |
[detail] |
820111 |
| 8.6 |
AT5G15740 |
O-fucosyltransferase family protein |
[detail] |
831430 |
| 8.4 |
BAG1 |
BCL-2-associated athanogene 1 |
[detail] |
835281 |
| 8.3 |
AT3G26950 |
uncharacterized protein |
[detail] |
822313 |
| 7.9 |
ATCSLA09 |
Nucleotide-diphospho-sugar transferases superfamily protein |
[detail] |
831734 |
| 7.6 |
GAE6 |
UDP-D-glucuronate 4-epimerase 6 |
[detail] |
821965 |
| 7.3 |
TBL28 |
TRICHOME BIREFRINGENCE-LIKE 28 |
[detail] |
818606 |
| 6.9 |
AT3G02250 |
O-fucosyltransferase family protein |
[detail] |
821223 |
| 6.8 |
DEAR3 |
DREB and EAR motif protein 3 |
[detail] |
816866 |
| 6.7 |
AT5G01790 |
uncharacterized protein |
[detail] |
831868 |
| 6.5 |
AT1G56670 |
GDSL-like Lipase/Acylhydrolase superfamily protein |
[detail] |
842122 |
| 6.3 |
GATA7 |
GATA transcription factor 7 |
[detail] |
829781 |
| 6.2 |
GATL3 |
galacturonosyltransferase-like 3 |
[detail] |
837885 |
| 5.9 |
TRM21 |
methyl-coenzyme M reductase II subunit gamma, putative (DUF3741) |
[detail] |
834410 |
| 5.4 |
IQD23 |
IQ-domain 23 |
[detail] |
836327 |
| 5.1 |
RBL6 |
RHOMBOID-like protein 6 |
[detail] |
837831 |
| 5.0 |
tny |
Integrase-type DNA-binding superfamily protein |
[detail] |
832650 |
| 5.0 |
MES3 |
methyl esterase 3 |
[detail] |
816893 |
| 4.9 |
AT1G72500 |
inter alpha-trypsin inhibitor, heavy chain-like protein |
[detail] |
843582 |
| 4.8 |
AT5G19025 |
Ribosomal protein L34e superfamily protein |
[detail] |
10723085 |
| 4.6 |
NUDX1 |
nudix hydrolase 1 |
[detail] |
843207 |
| 4.5 |
AT4G21890 |
zinc finger MYND domain protein |
[detail] |
828278 |
|
Coexpressed
gene list |
[Coexpressed gene list for NDL2]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
250337_at
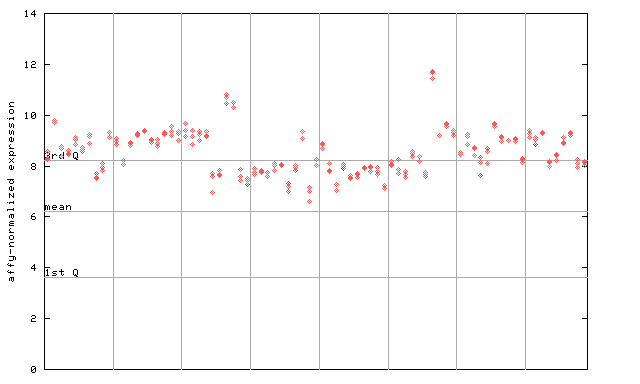
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
250337_at
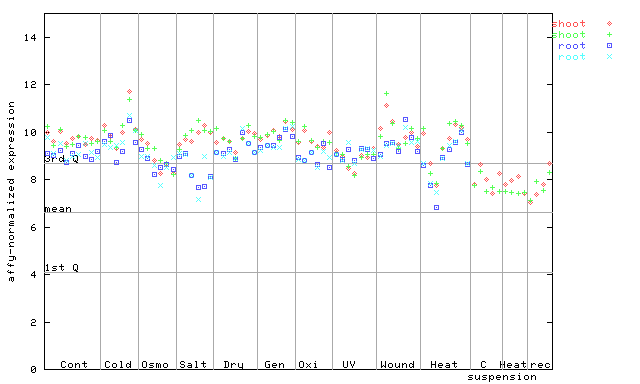
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
250337_at
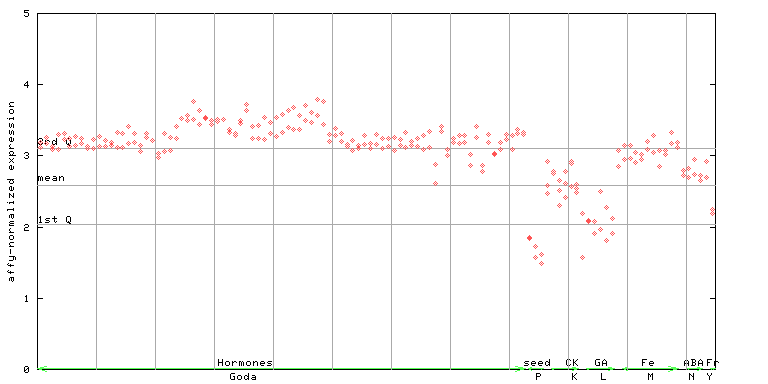
X axis is samples (xls file), and Y axis is log-expression.
|












