| functional annotation |
| Function |
exportin 1A |


|
| GO BP |
|
GO:0000056 [list] [network] ribosomal small subunit export from nucleus
|
(4 genes)
|
IBA
|
 |
|
GO:0046825 [list] [network] regulation of protein export from nucleus
|
(4 genes)
|
IBA
|
 |
|
GO:0000055 [list] [network] ribosomal large subunit export from nucleus
|
(5 genes)
|
IBA
|
 |
|
GO:0006611 [list] [network] protein export from nucleus
|
(45 genes)
|
ISS
|
 |
|
GO:0051028 [list] [network] mRNA transport
|
(63 genes)
|
IEA
|
 |
|
GO:0009846 [list] [network] pollen germination
|
(67 genes)
|
IGI
|
 |
|
GO:0009860 [list] [network] pollen tube growth
|
(129 genes)
|
IGI
|
 |
|
GO:0009553 [list] [network] embryo sac development
|
(146 genes)
|
IGI
|
 |
|
GO:0009408 [list] [network] response to heat
|
(229 genes)
|
IMP
|
 |
|
GO:0009555 [list] [network] pollen development
|
(337 genes)
|
IGI
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0005049 [list] [network] nuclear export signal receptor activity
|
(6 genes)
|
IBA
|
 |
|
GO:0008536 [list] [network] Ran GTPase binding
|
(14 genes)
|
IEA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
ath03008 [list] [network] Ribosome biogenesis in eukaryotes (98 genes) |
 |
| ath03013 [list] [network] RNA transport (171 genes) |
 |
| Protein |
NP_001190324.1
NP_197204.1
|
| BLAST |
NP_001190324.1
NP_197204.1
|
| Orthologous |
[Ortholog page]
XPO1B (ath)
LOC4334849 (osa)
LOC11435528 (mtr)
LOC11437746 (mtr)
LOC100256288 (vvi)
Xpo1 (sly)
LOC100384227 (zma)
LOC100783433 (gma)
LOC100787912 (gma)
LOC100802384 (gma)
LOC100813443 (gma)
LOC103625708 (zma)
LOC103844050 (bra)
LOC103846276 (bra)
LOC103851080 (bra)
LOC103856228 (bra)
|
Subcellular
localization
wolf |
|
nucl 2,
chlo 2,
extr 2,
cysk_nucl 1,
cyto 1,
mito 1,
plas 1,
vacu 1,
mito_plas 1,
cyto_plas 1
|
(predict for NP_001190324.1)
|
|
nucl 2,
chlo 2,
extr 2,
cysk_nucl 1,
cyto 1,
mito 1,
plas 1,
vacu 1,
mito_plas 1,
cyto_plas 1
|
(predict for NP_197204.1)
|
|
Subcellular
localization
TargetP |
|
other 8
|
(predict for NP_001190324.1)
|
|
other 8
|
(predict for NP_197204.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03018 |
RNA degradation |
4 |
 |
| ath03008 |
Ribosome biogenesis in eukaryotes |
3 |
 |
| ath03013 |
RNA transport |
3 |
 |
| ath00970 |
Aminoacyl-tRNA biosynthesis |
2 |
 |
Genes directly connected with XPO1A on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 9.5 |
XPO1B |
exportin 1B |
[detail] |
821074 |
| 8.3 |
GSL8 |
glucan synthase-like 8 |
[detail] |
818258 |
| 8.2 |
AT4G10320 |
tRNA synthetase class I (I, L, M and V) family protein |
[detail] |
826624 |
| 7.3 |
AT5G35430 |
Tetratricopeptide repeat (TPR)-like superfamily protein |
[detail] |
833507 |
| 6.9 |
AT3G61240 |
DEA(D/H)-box RNA helicase family protein |
[detail] |
825296 |
| 6.7 |
RH8 |
RNAhelicase-like 8 |
[detail] |
828042 |
| 6.7 |
AT4G15020 |
hAT transposon superfamily |
[detail] |
827161 |
|
Coexpressed
gene list |
[Coexpressed gene list for XPO1A]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
246424_at
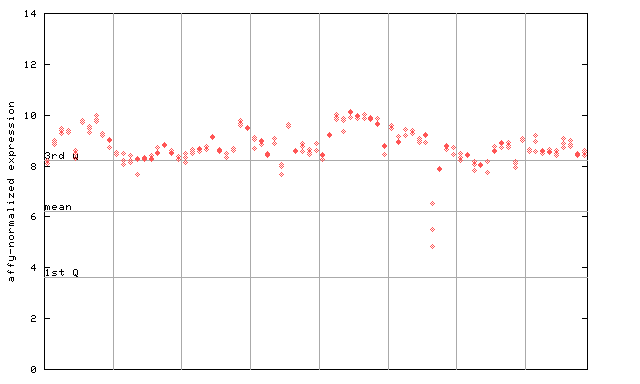
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
246424_at
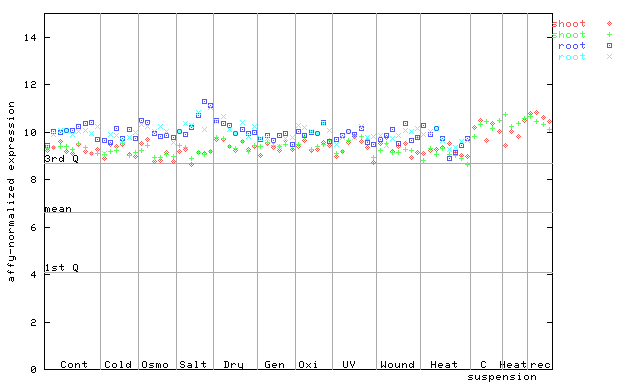
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
246424_at
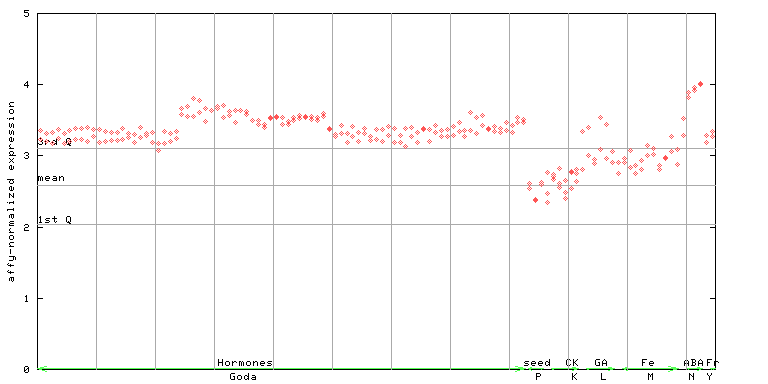
X axis is samples (xls file), and Y axis is log-expression.
|
















