| functional annotation |
| Function |
MEI2-like protein 1 |


|
| GO BP |
|
GO:0045836 [list] [network] positive regulation of meiotic nuclear division
|
(6 genes)
|
IGI
|
 |
|
GO:0000381 [list] [network] regulation of alternative mRNA splicing, via spliceosome
|
(30 genes)
|
IBA
|
 |
|
GO:0045292 [list] [network] mRNA cis splicing, via spliceosome
|
(38 genes)
|
IBA
|
 |
|
GO:0051321 [list] [network] meiotic cell cycle
|
(166 genes)
|
IEA
|
 |
|
GO:0048507 [list] [network] meristem development
|
(207 genes)
|
IGI
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001032122.1
NP_001318861.1
NP_001330764.1
NP_001330765.1
NP_001330766.1
NP_001330767.1
NP_001330768.1
NP_001330769.1
NP_001330770.1
NP_001330771.1
NP_568946.1
|
| BLAST |
NP_001032122.1
NP_001318861.1
NP_001330764.1
NP_001330765.1
NP_001330766.1
NP_001330767.1
NP_001330768.1
NP_001330769.1
NP_001330770.1
NP_001330771.1
NP_568946.1
|
| Orthologous |
[Ortholog page]
AML1 (sly)
ML4 (ath)
LOC7456646 (ppo)
LOC7481178 (ppo)
LOC9271988 (osa)
LOC11427537 (mtr)
LOC11429692 (mtr)
LOC11443824 (mtr)
LOC100192762 (zma)
LOC100193681 (zma)
LOC100260954 (vvi)
LOC100261030 (vvi)
LOC100797616 (gma)
LOC100797697 (gma)
LOC100797893 (gma)
LOC100801025 (gma)
LOC103653544 (zma)
LOC103655045 (zma)
LOC103847071 (bra)
LOC103854965 (bra)
LOC103874632 (bra)
LOC106798747 (gma)
|
Subcellular
localization
wolf |
|
nucl 7,
pero 1,
extr 1,
vacu 1,
cyto_pero 1
|
(predict for NP_001032122.1)
|
|
nucl 7,
pero 1,
extr 1,
vacu 1,
cyto_pero 1
|
(predict for NP_001318861.1)
|
|
nucl 8,
pero 1
|
(predict for NP_001330764.1)
|
|
nucl 7,
pero 1,
extr 1,
vacu 1,
cyto_pero 1
|
(predict for NP_001330765.1)
|
|
nucl 8,
pero 1
|
(predict for NP_001330766.1)
|
|
nucl 8,
pero 1
|
(predict for NP_001330767.1)
|
|
nucl 8,
pero 1
|
(predict for NP_001330768.1)
|
|
nucl 8,
pero 1
|
(predict for NP_001330769.1)
|
|
nucl 8,
pero 1
|
(predict for NP_001330770.1)
|
|
nucl 8,
pero 1
|
(predict for NP_001330771.1)
|
|
nucl 7,
pero 1,
extr 1,
vacu 1,
cyto_pero 1
|
(predict for NP_568946.1)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_001032122.1)
|
|
other 9
|
(predict for NP_001318861.1)
|
|
other 8
|
(predict for NP_001330764.1)
|
|
other 9
|
(predict for NP_001330765.1)
|
|
other 8
|
(predict for NP_001330766.1)
|
|
other 8
|
(predict for NP_001330767.1)
|
|
other 8
|
(predict for NP_001330768.1)
|
|
other 8
|
(predict for NP_001330769.1)
|
|
other 8
|
(predict for NP_001330770.1)
|
|
other 8
|
(predict for NP_001330771.1)
|
|
other 9
|
(predict for NP_568946.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03018 |
RNA degradation |
3 |
 |
Genes directly connected with ML1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 9.0 |
ML4 |
MEI2-like 4 |
[detail] |
830620 |
| 7.4 |
UBP25 |
ubiquitin-specific protease 25 |
[detail] |
820662 |
| 6.9 |
AT5G60170 |
RNA binding (RRM/RBD/RNP motifs) family protein |
[detail] |
836139 |
| 6.3 |
OXS2 |
CCCH-type zinc finger protein with ARM repeat domain-containing protein |
[detail] |
818790 |
| 5.7 |
Phox3 |
Octicosapeptide/Phox/Bem1p (PB1) domain-containing protein / tetratricopeptide repeat (TPR)-containing protein |
[detail] |
832158 |
|
Coexpressed
gene list |
[Coexpressed gene list for ML1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
247506_at
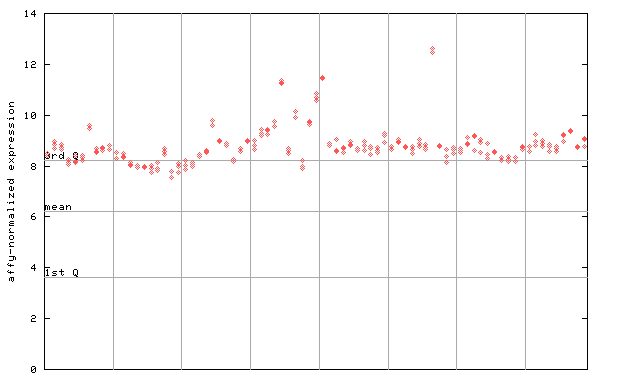
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
247506_at
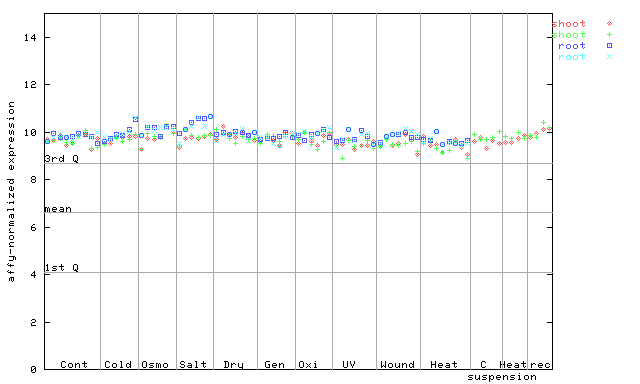
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
247506_at
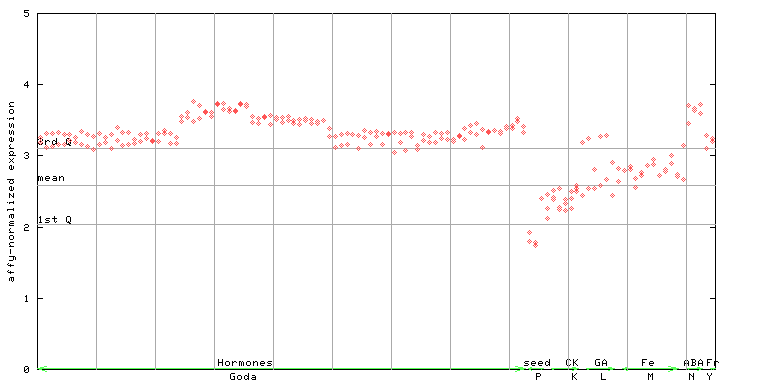
X axis is samples (xls file), and Y axis is log-expression.
|







