| functional annotation |
| Function |
vacuolar protein sorting 34 |


|
| GO BP |
|
GO:0036092 [list] [network] phosphatidylinositol-3-phosphate biosynthetic process
|
(2 genes)
|
IBA
|
 |
|
GO:0030242 [list] [network] autophagy of peroxisome
|
(3 genes)
|
IBA
|
 |
|
GO:0048015 [list] [network] phosphatidylinositol-mediated signaling
|
(16 genes)
|
IBA
|
 |
|
GO:0000045 [list] [network] autophagosome assembly
|
(19 genes)
|
IBA
|
 |
|
GO:0046854 [list] [network] phosphatidylinositol phosphorylation
|
(24 genes)
|
IBA
|
 |
|
GO:0055046 [list] [network] microgametogenesis
|
(55 genes)
|
IMP
|
 |
|
GO:0006897 [list] [network] endocytosis
|
(60 genes)
|
IBA
IDA
IMP
|
 |
|
GO:0072593 [list] [network] reactive oxygen species metabolic process
|
(133 genes)
|
IMP
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(485 genes)
|
IMP
|
 |
|
| GO CC |
|
GO:0034271 [list] [network] phosphatidylinositol 3-kinase complex, class III, type I
|
(3 genes)
|
IBA
|
 |
|
GO:0034272 [list] [network] phosphatidylinositol 3-kinase complex, class III, type II
|
(3 genes)
|
IBA
|
 |
|
GO:0000407 [list] [network] phagophore assembly site
|
(15 genes)
|
IBA
|
 |
|
GO:0005777 [list] [network] peroxisome
|
(325 genes)
|
IBA
|
 |
|
GO:0005768 [list] [network] endosome
|
(424 genes)
|
IBA
|
 |
|
GO:0005794 [list] [network] Golgi apparatus
|
(1430 genes)
|
ISM
|
 |
|
GO:0016020 [list] [network] membrane
|
(7839 genes)
|
IBA
|
 |
|
GO:0005737 [list] [network] cytoplasm
|
(14855 genes)
|
IBA
|
 |
|
| GO MF |
|
GO:0016303 [list] [network] 1-phosphatidylinositol-3-kinase activity
|
(2 genes)
|
IBA
|
 |
|
GO:0005524 [list] [network] ATP binding
|
(2003 genes)
|
IEA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
ath00562 [list] [network] Inositol phosphate metabolism (77 genes) |
 |
| ath04070 [list] [network] Phosphatidylinositol signaling system (76 genes) |
 |
| ath04136 [list] [network] Autophagy - other (40 genes) |
 |
| ath04145 [list] [network] Phagosome (82 genes) |
 |
| Protein |
NP_176251.1
|
| BLAST |
NP_176251.1
|
| Orthologous |
[Ortholog page]
LOC547983 (gma)
LOC4337982 (osa)
LOC7484773 (ppo)
LOC11415599 (mtr)
LOC100263708 (vvi)
LOC100778348 (gma)
LOC101250591 (sly)
LOC103653397 (zma)
LOC103838506 (bra)
|
Subcellular
localization
wolf |
|
nucl 4,
cyto 4,
cyto_nucl 4
|
(predict for NP_176251.1)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_176251.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04141 |
Protein processing in endoplasmic reticulum |
4 |
 |
| ath04144 |
Endocytosis |
3 |
 |
| ath04145 |
Phagosome |
2 |
 |
| ath04120 |
Ubiquitin mediated proteolysis |
2 |
 |
| ath03050 |
Proteasome |
2 |
 |
Genes directly connected with VPS34 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.3 |
SKD1 |
AAA-type ATPase family protein |
[detail] |
817306 |
| 6.1 |
AT1G69340 |
appr-1-p processing enzyme family protein |
[detail] |
843266 |
| 6.1 |
AT2G17190 |
ubiquitin family protein |
[detail] |
816224 |
| 5.3 |
FAAH |
fatty acid amide hydrolase |
[detail] |
836565 |
|
Coexpressed
gene list |
[Coexpressed gene list for VPS34]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
264927_at
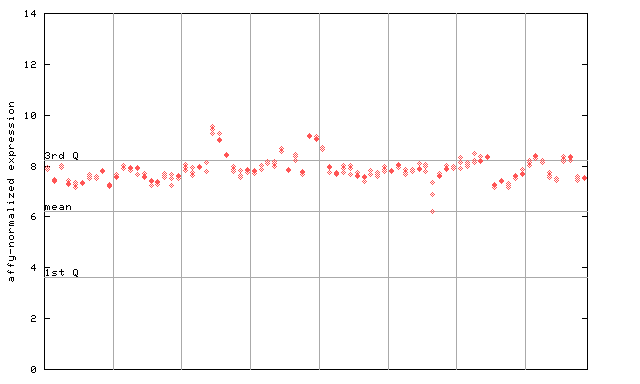
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
264927_at
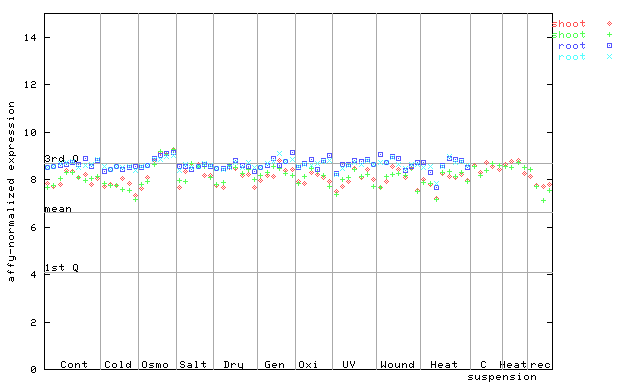
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
264927_at
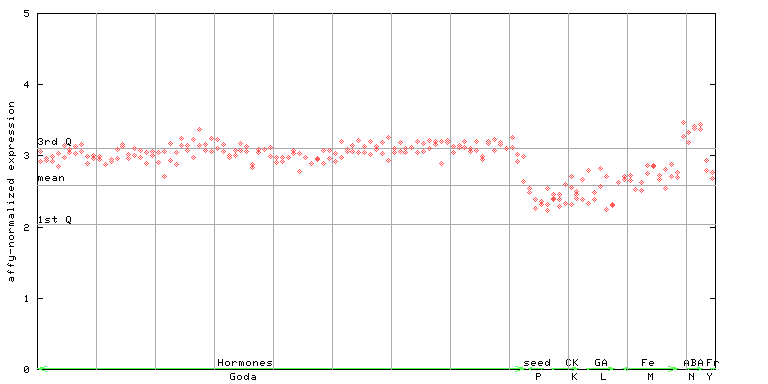
X axis is samples (xls file), and Y axis is log-expression.
|


















