[←][→] ath
| functional annotation | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | branched-chain amino acid transaminase 2 |


|
|||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | ath00270 [list] [network] Cysteine and methionine metabolism (121 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||
| ath00280 [list] [network] Valine, leucine and isoleucine degradation (51 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00290 [list] [network] Valine, leucine and isoleucine biosynthesis (22 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00770 [list] [network] Pantothenate and CoA biosynthesis (28 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00966 [list] [network] Glucosinolate biosynthesis (23 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||
| ath01210 [list] [network] 2-Oxocarboxylic acid metabolism (74 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||
| ath01230 [list] [network] Biosynthesis of amino acids (251 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_001031015.1 NP_001031016.2 NP_172478.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_001031015.1 NP_001031016.2 NP_172478.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] LOC4332151 (osa) LOC7467241 (ppo) LOC25490006 (mtr) LOC100260903 (vvi) LOC100266106 (vvi) LOC100281255 (zma) LOC100782644 (gma) LOC100818776 (gma) LOC101253875 (sly) LOC101254608 (sly) LOC103843316 (bra) LOC103868121 (bra) LOC103871787 (bra) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for BCAT-2] | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | |||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | ||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
264524_at
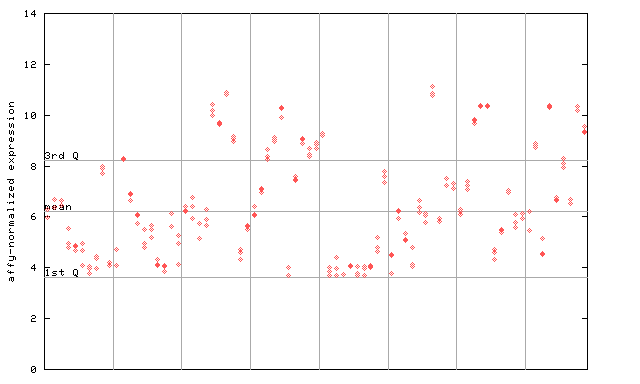
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
264524_at
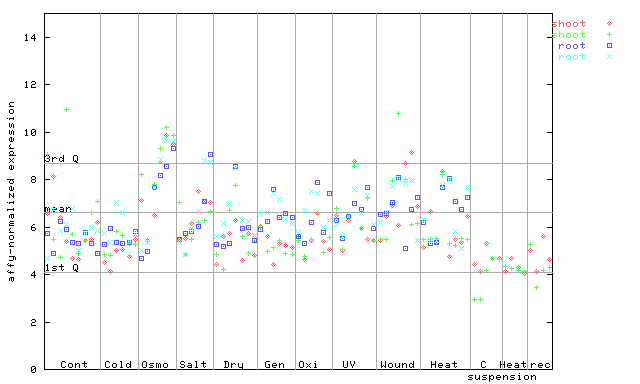
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
264524_at
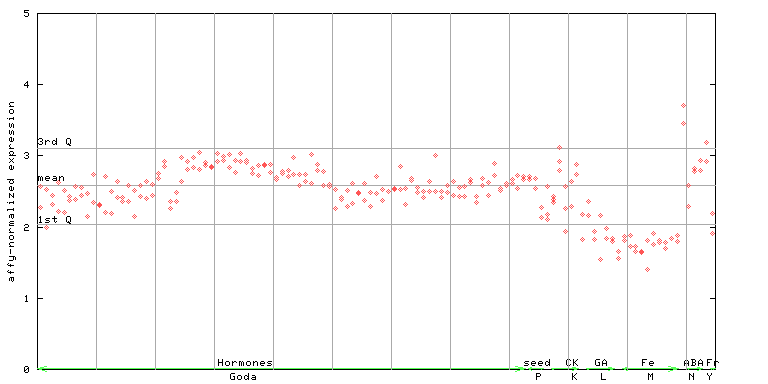
X axis is samples (xls file), and Y axis is log-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 837543 |


|
| Refseq ID (protein) | NP_001031015.1 |  |
| NP_001031016.2 |  |
|
| NP_172478.1 |  |
|
The preparation time of this page was 0.2 [sec].





