| functional annotation |
| Function |
NAC domain containing protein 70 |

 Plant GARDEN Plant GARDEN JBrowse
Plant GARDEN Plant GARDEN JBrowse
|
| GO BP |
|
GO:0048829 [list] [network] root cap development
|
(16 genes)
|
IMP
|
 |
|
GO:0010628 [list] [network] positive regulation of gene expression
|
(71 genes)
|
IEP
|
 |
|
GO:0009834 [list] [network] plant-type secondary cell wall biogenesis
|
(97 genes)
|
IMP
|
 |
|
GO:0045893 [list] [network] positive regulation of DNA-templated transcription
|
(272 genes)
|
IMP
|
 |
|
GO:0006355 [list] [network] regulation of DNA-templated transcription
|
(1656 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0003677 [list] [network] DNA binding
|
(1277 genes)
|
IEA
|
 |
|
GO:0003700 [list] [network] DNA-binding transcription factor activity
|
(1576 genes)
|
ISS
|
 |
|
| KEGG |
|
|
| Protein |
NP_001329933.1
NP_192773.1
|
| BLAST |
NP_001329933.1
NP_192773.1
|
| Orthologous |
[Ortholog page]
NAC015 (ath)
LOC7479032 (ppo)
LOC11434735 (mtr)
LOC18108387 (ppo)
LOC100775746 (gma)
NAC171 (gma)
LOC101261423 (sly)
LOC103838908 (bra)
LOC103858562 (bra)
LOC107280688 (osa)
LOC123041433 (tae)
LOC123049420 (tae)
LOC123185513 (tae)
LOC123430320 (hvu)
|
Subcellular
localization
wolf |
|
nucl 7,
chlo 1,
cyto 1,
mito 1,
plas 1,
chlo_mito 1,
mito_plas 1,
cyto_plas 1
|
(predict for NP_001329933.1)
|
|
nucl 7,
pero 2
|
(predict for NP_192773.1)
|
|
Subcellular
localization
TargetP |
|
other 7
|
(predict for NP_001329933.1)
|
|
other 8
|
(predict for NP_192773.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with NAC070 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 15.9 |
NAC015 |
NAC domain containing protein 15 |
[detail] |
840223 |
| 12.0 |
AT5G24070 |
Peroxidase superfamily protein |
[detail] |
832472 |
| 11.8 |
sks16 |
SKU5 similar 16 |
[detail] |
816895 |
| 11.4 |
AT1G16905 |
Curculin-like (mannose-binding) lectin family protein |
[detail] |
5007699 |
| 11.1 |
AT5G51520 |
Plant invertase/pectin methylesterase inhibitor superfamily protein |
[detail] |
835226 |
| 11.0 |
AT4G01240 |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
[detail] |
827909 |
| 10.9 |
AT1G65570 |
Pectin lyase-like superfamily protein |
[detail] |
842868 |
| 10.6 |
AT5G58784 |
Undecaprenyl pyrophosphate synthetase family protein |
[detail] |
835994 |
| 9.1 |
AT1G05280 |
ERV-F (C)1 provirus ancestral Env polyprotein, putative (DUF604) |
[detail] |
837026 |
| 8.6 |
AT5G20045 |
uncharacterized protein |
[detail] |
2745992 |
| 8.5 |
SMB |
NAC (No Apical Meristem) domain transcriptional regulator superfamily protein |
[detail] |
844296 |
| 6.2 |
AT4G23590 |
Tyrosine transaminase family protein |
[detail] |
828459 |
| 6.1 |
LBD15 |
LOB domain-containing protein 15 |
[detail] |
818641 |
| 5.6 |
AT1G73340 |
Cytochrome P450 superfamily protein |
[detail] |
843669 |
| 4.8 |
RGXT1 |
rhamnogalacturonan xylosyltransferase 1 |
[detail] |
828129 |
|
Coexpressed
gene list |
[Coexpressed gene list for NAC070]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
255022_at
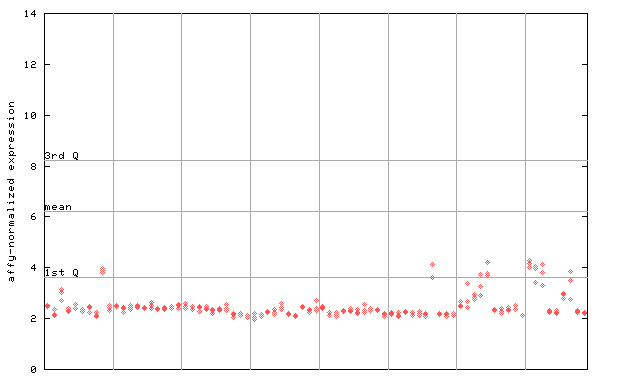
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
255022_at
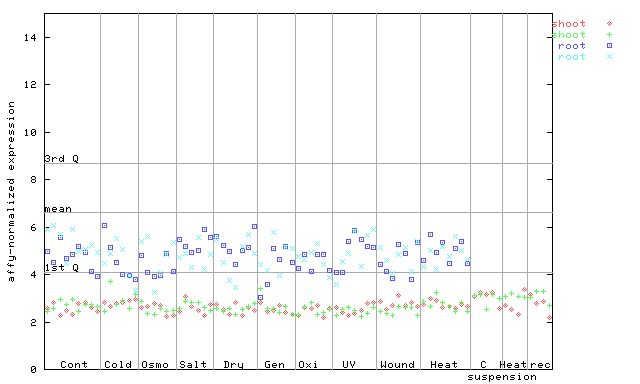
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
255022_at
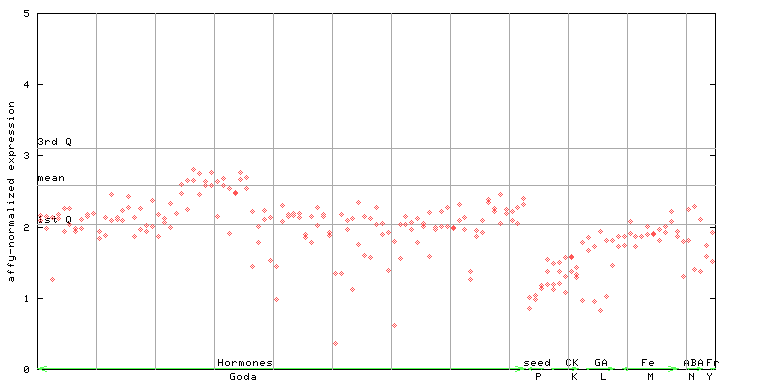
X axis is samples (xls file), and Y axis is log-expression.
|

 Plant GARDEN Plant GARDEN JBrowse
Plant GARDEN Plant GARDEN JBrowse








