| functional annotation |
| Function |
Protein kinase superfamily protein |




|
| GO BP |
|
GO:1902171 [list] [network] regulation of tocopherol cyclase activity
|
(2 genes)
|
IDA
|
 |
|
GO:0080177 [list] [network] plastoglobule organization
|
(3 genes)
|
IMP
|
 |
|
GO:1904143 [list] [network] positive regulation of carotenoid biosynthetic process
|
(4 genes)
|
IMP
|
 |
|
GO:1901562 [list] [network] response to paraquat
|
(5 genes)
|
IEP
|
 |
|
GO:1902326 [list] [network] positive regulation of chlorophyll biosynthetic process
|
(7 genes)
|
IMP
|
 |
|
GO:0080183 [list] [network] response to photooxidative stress
|
(13 genes)
|
IMP
|
 |
|
GO:0015996 [list] [network] chlorophyll catabolic process
|
(18 genes)
|
IMP
|
 |
|
GO:0031540 [list] [network] regulation of anthocyanin biosynthetic process
|
(18 genes)
|
IMP
|
 |
|
GO:0006995 [list] [network] cellular response to nitrogen starvation
|
(27 genes)
|
IMP
|
 |
|
GO:0009767 [list] [network] photosynthetic electron transport chain
|
(39 genes)
|
IMP
|
 |
|
GO:0010109 [list] [network] regulation of photosynthesis
|
(47 genes)
|
IMP
|
 |
|
GO:0010027 [list] [network] thylakoid membrane organization
|
(49 genes)
|
IMP
|
 |
|
GO:0010114 [list] [network] response to red light
|
(101 genes)
|
IMP
|
 |
|
GO:0009637 [list] [network] response to blue light
|
(160 genes)
|
IMP
|
 |
|
GO:0006468 [list] [network] protein phosphorylation
|
(639 genes)
|
IEA
|
 |
|
GO:0009414 [list] [network] response to water deprivation
|
(1006 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001031763.1
NP_001329437.1
NP_194867.2
|
| BLAST |
NP_001031763.1
NP_001329437.1
NP_194867.2
|
| Orthologous |
[Ortholog page]
LOC4350081 (osa)
LOC18100763 (ppo)
LOC25481877 (mtr)
LOC100790292 (gma)
LOC100819754 (gma)
ABC1K1 (sly)
LOC103851895 (bra)
LOC123086550 (tae)
LOC123091394 (tae)
LOC123096362 (tae)
LOC123447878 (hvu)
|
Subcellular
localization
wolf |
|
chlo 6,
chlo_mito 4,
mito 2
|
(predict for NP_001031763.1)
|
|
cyto 7,
chlo 1,
nucl 1,
cysk 1,
chlo_mito 1,
cysk_nucl 1
|
(predict for NP_001329437.1)
|
|
chlo 6,
chlo_mito 4,
mito 2
|
(predict for NP_194867.2)
|
|
Subcellular
localization
TargetP |
|
chlo 9
|
(predict for NP_001031763.1)
|
|
other 8
|
(predict for NP_001329437.1)
|
|
chlo 9
|
(predict for NP_194867.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00906 |
Carotenoid biosynthesis |
2 |
 |
Genes directly connected with ACDO1 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 8.3 |
SVR3 |
elongation factor family protein |
[detail] |
831209 |
| 7.5 |
AT3G24190 |
Protein kinase superfamily protein |
[detail] |
822005 |
| 7.4 |
ZDS |
zeta-carotene desaturase |
[detail] |
819647 |
| 7.3 |
F2KP |
fructose-2,6-bisphosphatase |
[detail] |
837221 |
| 6.9 |
ATSPS4F |
Sucrose-phosphate synthase family protein |
[detail] |
826603 |
| 6.8 |
AT5G57960 |
GTP-binding protein, HflX |
[detail] |
835907 |
|
Coexpressed
gene list |
[Coexpressed gene list for ACDO1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
253517_at
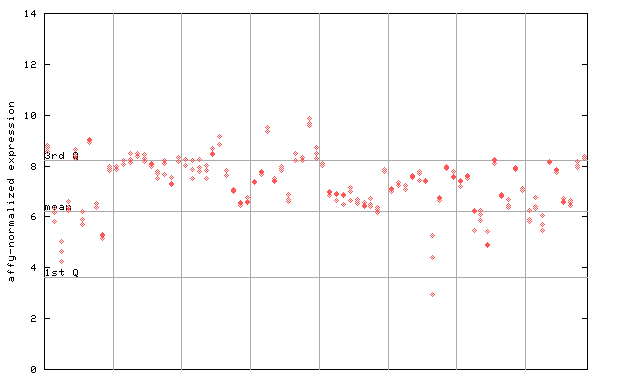
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
253517_at
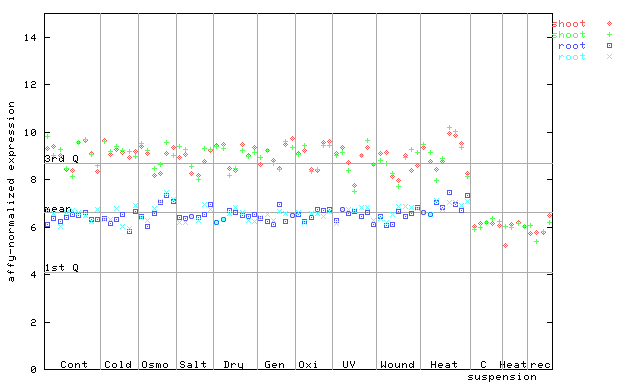
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
253517_at
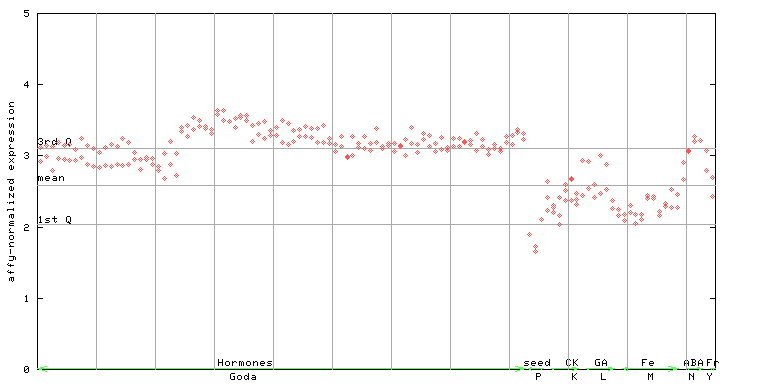
X axis is samples (xls file), and Y axis is log-expression.
|














