| functional annotation |
| Function |
RNA polymerase II large subunit |


|
| GO BP |
|
GO:0006366 [list] [network] transcription by RNA polymerase II
|
(11 genes)
|
IEA
|
 |
|
GO:0060966 [list] [network] regulation of gene silencing by RNA
|
(25 genes)
|
IEA
|
 |
|
GO:0070918 [list] [network] regulatory ncRNA processing
|
(51 genes)
|
IEA
|
 |
|
GO:0010628 [list] [network] positive regulation of gene expression
|
(71 genes)
|
IEA
|
 |
|
GO:0035194 [list] [network] RNA-mediated post-transcriptional gene silencing
|
(92 genes)
|
IEA
|
 |
|
GO:0048573 [list] [network] photoperiodism, flowering
|
(94 genes)
|
IEA
|
 |
|
GO:0040029 [list] [network] epigenetic regulation of gene expression
|
(146 genes)
|
IEA
|
 |
|
GO:0043414 [list] [network] macromolecule methylation
|
(153 genes)
|
IEA
|
 |
|
GO:0008380 [list] [network] RNA splicing
|
(191 genes)
|
IEA
|
 |
|
GO:0045892 [list] [network] negative regulation of DNA-templated transcription
|
(205 genes)
|
IEA
|
 |
|
GO:0006259 [list] [network] DNA metabolic process
|
(381 genes)
|
IEA
|
 |
|
GO:0048507 [list] [network] meristem development
|
(457 genes)
|
IEA
|
 |
|
GO:2000241 [list] [network] regulation of reproductive process
|
(507 genes)
|
IEA
|
 |
|
GO:0009793 [list] [network] embryo development ending in seed dormancy
|
(697 genes)
|
IEA
|
 |
|
GO:0048522 [list] [network] positive regulation of cellular process
|
(856 genes)
|
IEA
|
 |
|
GO:0048589 [list] [network] developmental growth
|
(1060 genes)
|
IEA
|
 |
|
GO:0050793 [list] [network] regulation of developmental process
|
(1250 genes)
|
IEA
|
 |
|
GO:0048367 [list] [network] shoot system development
|
(1318 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
ath03020 [list] [network] RNA polymerase (52 genes) |
 |
| Protein |
NP_001329346.1
NP_195305.2
|
| BLAST |
NP_001329346.1
NP_195305.2
|
| Orthologous |
[Ortholog page]
LOC778257 (sly)
LOC4337831 (osa)
LOC9267835 (osa)
LOC11434004 (mtr)
LOC18099128 (ppo)
LOC100778777 (gma)
LOC100804975 (gma)
LOC100812506 (gma)
LOC103834927 (bra)
LOC103873389 (bra)
LOC123047603 (tae)
LOC123114040 (tae)
LOC123114042 (tae)
LOC123123534 (tae)
LOC123123536 (tae)
LOC123147362 (tae)
LOC123155625 (tae)
LOC123171586 (tae)
LOC123191235 (tae)
LOC123397332 (hvu)
LOC123407901 (hvu)
LOC123428554 (hvu)
|
Subcellular
localization
wolf |
|
nucl 8,
cyto 1,
mito 1,
cysk 1
|
(predict for NP_001329346.1)
|
|
nucl 8,
cyto 1,
mito 1,
cysk 1
|
(predict for NP_195305.2)
|
|
Subcellular
localization
TargetP |
|
other 6
|
(predict for NP_001329346.1)
|
|
other 6
|
(predict for NP_195305.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03040 |
Spliceosome |
2 |
 |
Genes directly connected with NRPB1 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 12.6 |
AT3G50370 |
uncharacterized protein |
[detail] |
824199 |
| 12.1 |
CHR4 |
chromatin remodeling 4 |
[detail] |
834510 |
| 11.8 |
SUS2 |
Pre-mRNA-processing-splicing factor |
[detail] |
844347 |
| 11.1 |
HUA2 |
Tudor/PWWP/MBT domain-containing protein |
[detail] |
832379 |
| 9.9 |
LBA1 |
RNA helicase |
[detail] |
834747 |
| 9.5 |
AT5G51300 |
splicing factor-like protein |
[detail] |
835204 |
| 7.7 |
AT5G27650 |
Tudor/PWWP/MBT superfamily protein |
[detail] |
832827 |
| 7.5 |
AT1G58220 |
Homeodomain-like superfamily protein |
[detail] |
842189 |
| 6.8 |
AT5G18230 |
transcription regulator NOT2/NOT3/NOT5 family protein |
[detail] |
831941 |
|
Coexpressed
gene list |
[Coexpressed gene list for NRPB1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
253133_at
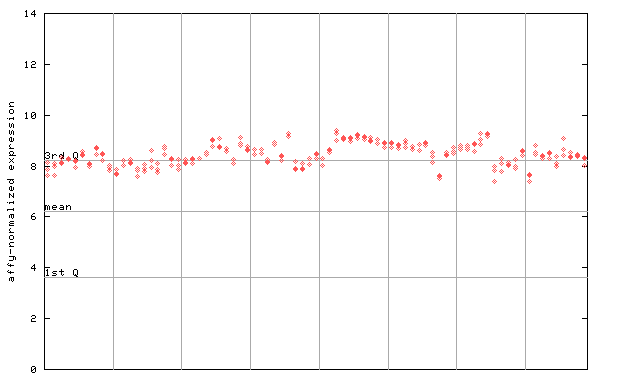
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
253133_at
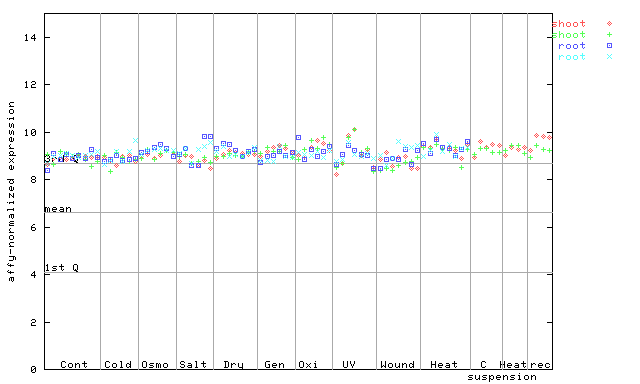
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
253133_at
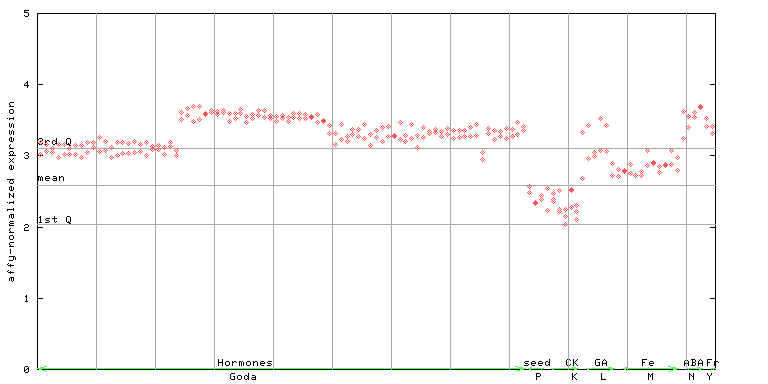
X axis is samples (xls file), and Y axis is log-expression.
|












