| functional annotation |
| Function |
plasma-membrane associated cation-binding protein 1 |




|
| GO BP |
|
GO:0010350 [list] [network] cellular response to magnesium starvation
|
(1 genes)
|
IEP
|
 |
|
GO:0071286 [list] [network] cellular response to magnesium ion
|
(1 genes)
|
IEP
|
 |
|
GO:0071325 [list] [network] cellular response to mannitol stimulus
|
(1 genes)
|
IEP
|
 |
|
GO:0072709 [list] [network] cellular response to sorbitol
|
(1 genes)
|
IEP
|
 |
|
GO:0031115 [list] [network] negative regulation of microtubule polymerization
|
(2 genes)
|
IDA
|
 |
|
GO:0031117 [list] [network] positive regulation of microtubule depolymerization
|
(2 genes)
|
IDA
|
 |
|
GO:0071281 [list] [network] cellular response to iron ion
|
(2 genes)
|
IEP
|
 |
|
GO:0075733 [list] [network] intracellular transport of virus
|
(2 genes)
|
IMP
|
 |
|
GO:0006499 [list] [network] N-terminal protein myristoylation
|
(3 genes)
|
IMP
|
 |
|
GO:0035865 [list] [network] cellular response to potassium ion
|
(3 genes)
|
IEP
|
 |
|
GO:0071280 [list] [network] cellular response to copper ion
|
(3 genes)
|
IEP
|
 |
|
GO:0051511 [list] [network] negative regulation of unidimensional cell growth
|
(4 genes)
|
IMP
|
 |
|
GO:0071219 [list] [network] cellular response to molecule of bacterial origin
|
(11 genes)
|
IEP
|
 |
|
GO:0090332 [list] [network] stomatal closure
|
(21 genes)
|
IMP
|
 |
|
GO:0051592 [list] [network] response to calcium ion
|
(27 genes)
|
IDA
|
 |
|
GO:0071472 [list] [network] cellular response to salt stress
|
(41 genes)
|
IEP
|
 |
|
GO:0043622 [list] [network] cortical microtubule organization
|
(43 genes)
|
IDA
IMP
|
 |
|
GO:0009409 [list] [network] response to cold
|
(472 genes)
|
IEP
|
 |
|
GO:0009414 [list] [network] response to water deprivation
|
(1006 genes)
|
IEP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0005547 [list] [network] phosphatidylinositol-3,4,5-trisphosphate binding
|
(3 genes)
|
IDA
|
 |
|
GO:0043325 [list] [network] phosphatidylinositol-3,4-bisphosphate binding
|
(3 genes)
|
IDA
|
 |
|
GO:0080025 [list] [network] phosphatidylinositol-3,5-bisphosphate binding
|
(3 genes)
|
IDA
|
 |
|
GO:0005546 [list] [network] phosphatidylinositol-4,5-bisphosphate binding
|
(30 genes)
|
IDA
|
 |
|
GO:0008017 [list] [network] microtubule binding
|
(81 genes)
|
IDA
|
 |
|
GO:0005516 [list] [network] calmodulin binding
|
(128 genes)
|
IDA
|
 |
|
GO:0005507 [list] [network] copper ion binding
|
(132 genes)
|
IDA
|
 |
|
GO:0005509 [list] [network] calcium ion binding
|
(132 genes)
|
IDA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(5066 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001031676.1
NP_001031677.1
NP_001190778.1
NP_001190779.1
NP_001320005.1
NP_001329492.1
NP_001329493.1
NP_001329494.1
NP_193759.1
NP_849412.1
|
| BLAST |
NP_001031676.1
NP_001031677.1
NP_001190778.1
NP_001190779.1
NP_001320005.1
NP_001329492.1
NP_001329493.1
NP_001329494.1
NP_193759.1
NP_849412.1
|
| Orthologous |
[Ortholog page]
LOC4324284 (osa)
LOC4329036 (osa)
LOC7491458 (ppo)
LOC18097095 (ppo)
LOC25488186 (mtr)
LOC100499895 (gma)
LOC100786734 (gma)
LOC101260974 (sly)
LOC103827887 (bra)
LOC103839170 (bra)
LOC103858049 (bra)
LOC103861191 (bra)
LOC123060814 (tae)
LOC123069376 (tae)
LOC123077887 (tae)
LOC123443270 (hvu)
|
Subcellular
localization
wolf |
|
nucl 6,
cyto 4
|
(predict for NP_001031676.1)
|
|
nucl 6,
cyto 4
|
(predict for NP_001031677.1)
|
|
nucl 8,
cyto 1
|
(predict for NP_001190778.1)
|
|
nucl 6,
cyto 3
|
(predict for NP_001190779.1)
|
|
nucl 6,
cyto 4
|
(predict for NP_001320005.1)
|
|
nucl 6,
cyto 4
|
(predict for NP_001329492.1)
|
|
nucl 6,
cyto 4
|
(predict for NP_001329493.1)
|
|
nucl 6,
cyto 4
|
(predict for NP_001329494.1)
|
|
nucl 6,
cyto 4
|
(predict for NP_193759.1)
|
|
nucl 6,
cyto 4
|
(predict for NP_849412.1)
|
|
Subcellular
localization
TargetP |
|
other 6
|
(predict for NP_001031676.1)
|
|
other 5,
scret 3
|
(predict for NP_001031677.1)
|
|
other 6
|
(predict for NP_001190778.1)
|
|
other 6
|
(predict for NP_001190779.1)
|
|
other 6
|
(predict for NP_001320005.1)
|
|
other 6
|
(predict for NP_001329492.1)
|
|
other 6
|
(predict for NP_001329493.1)
|
|
other 6
|
(predict for NP_001329494.1)
|
|
other 6
|
(predict for NP_193759.1)
|
|
other 6
|
(predict for NP_849412.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04144 |
Endocytosis |
3 |
 |
| ath04145 |
Phagosome |
2 |
 |
|
Coexpressed
gene list |
[Coexpressed gene list for PCAP1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
254492_at
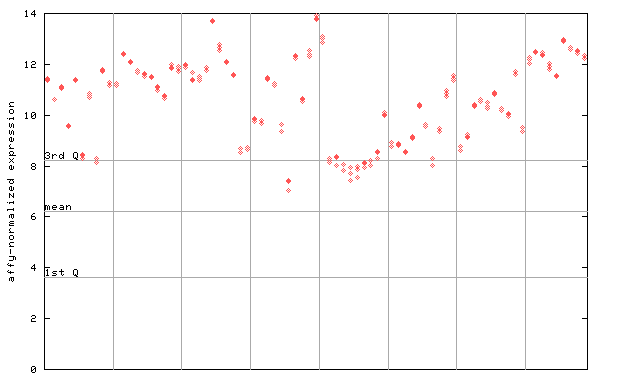
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
254492_at
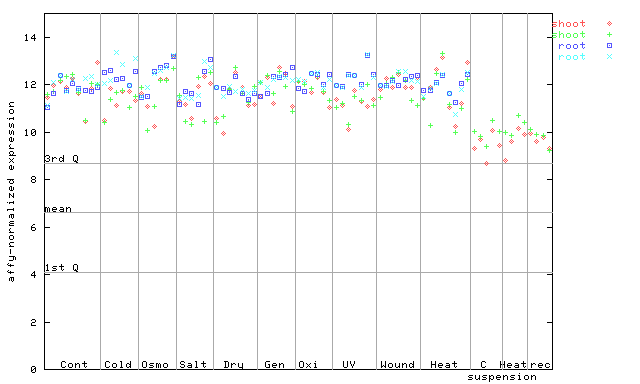
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
254492_at
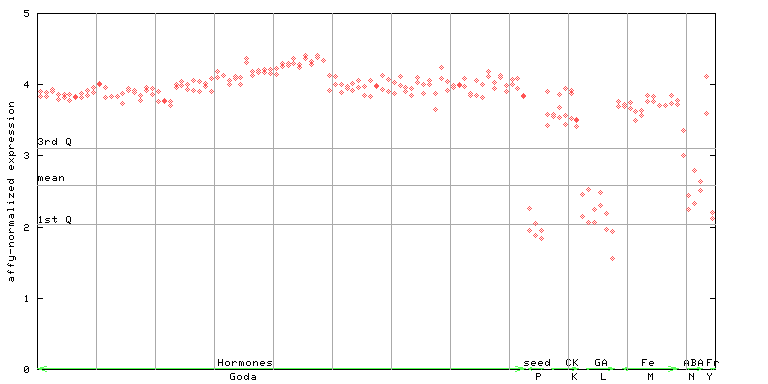
X axis is samples (xls file), and Y axis is log-expression.
|










