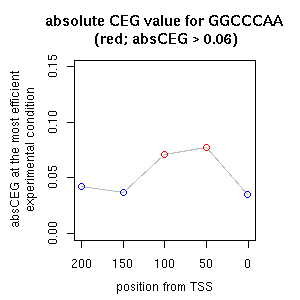
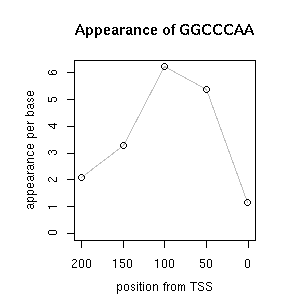
| reported seq | reported name | reference |
|---|---|---|
| GGCCCAWWW | UP1ATMSD | [ref] |
 |
 |
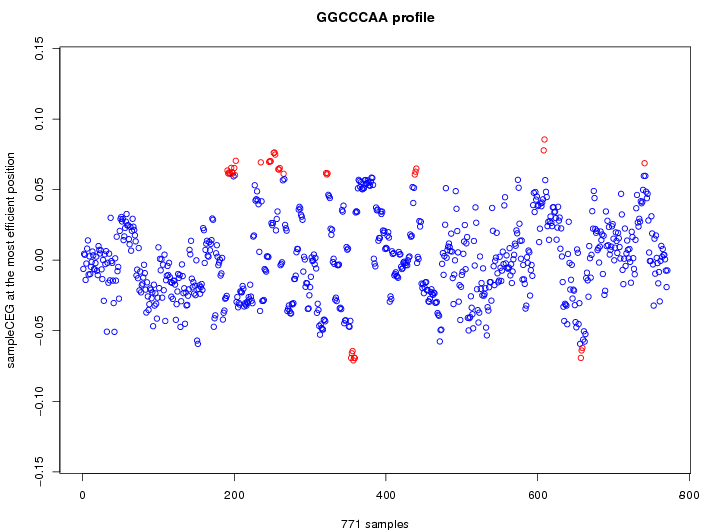
| GO ID | term | -Log(p) | number of {A; genes in a GO} | numver of {B; genes with this heptamer} | {A} x {B} |
|---|---|---|---|---|---|
| no functional bias detected | |||||
| correl | TF | function ((alias))* | TF family (link to AGRIS) |
|---|---|---|---|
| 0.70 | At1g71260 | expressed protein | WHIRLY |
| 0.68 | At4g30930 | 50S ribosomal protein L21, mitochondrial (RPL21M) ((WRKY32)) | WRKY |
| 0.62 | At1g14410 | DNA-binding protein-related | WHIRLY |
| 0.60 | At3g52910 | expressed protein | GRF |
| 0.60 | At3g57150 | dyskerin, putative / nucleolar protein NAP57, putative ((NAP57)) | NAC |
| 0.60 | At5g16070 | chaperonin, putative | TCP |
| -0.59 | At1g08320 | bZIP family transcription factor | bZIP |
| -0.56 | At1g19310 | zinc finger (C3HC4-type RING finger) family protein | C3H |
| -0.56 | At3g02340 | zinc finger (C3HC4-type RING finger) family protein | C3H |
| -0.55 | At3g19580 | zinc finger (C2H2 type) protein 2 (AZF2) ((AZF2)) | C2H2 |
| -0.55 | At4g27410 | no apical meristem (NAM) family protein (RD26) ((RD26)) | NAC |
| -0.54 | At5g04340 | zinc finger (C2H2 type) family protein ((C2H2)) | C2H2 |
| 0.54 | At3g22780 | CXC domain protein (TSO1) ((TSO1)) | CPP |
| 0.53 | At2g02740 | transcription factor, putative | WHIRLY |
| -0.53 | At3g52800 | zinc finger (AN1-like) family protein | C2H2 |
|