Node annotations (aliases)
Under the AGI code in nodes, short functional annotation is displayed.
- When alias is given for a gene, the alias is used for this short functional annotation.
- '?' indicates 'expressed protein' or 'hypothetical protein'.
- The terms 'putative', 'related', 'family', '-containing', 'protein' and information of protein localization were deleted for this short annotation.
Node annotations (KEGG pathway)
Small marks (red, yellow, etc.) on nodes indicate a common KEGG pathway in the networks. For each gene network, up to five KEGG pathways with larger numbers of genes are selected.
Node shape
| Shape | Description |
|---|
| Octagon | Transcription factors |
| Circle | Others |
Annotations of transcription factor genes are derived from AGRIS.
Coexpressed gene networks in ATTED-II are drawn based on rank of correlation.
Use of correlation rank partially normalizes the density of genes in correlation space.
Correlation rank is asymmetric, namely the rank of gene B from gene A is not equal with the rank of gene A from gene B.
Thus, the geometric averaged rank is used.
MR(AB) = √ (Rank(A→B) x Rank(B→A))
| MR* | Line thickness |
|---|
| 1 ~ 5 | Bold lines |
| 5 ~ 30 | Normal lines |
| 30 ~ | Weak lines |
For every (reference) gene, the top 3 MR genes are connected to the reference gene.
Edge types
Edge types indecate supporting data for the gene pairs.
| Edge type | Description | Data source |
|---|
| Orange | Conserved coexpression | Conserved coexpressed is deduced from the comparison with mammalian coexpression data (Human, Mouse and Rat) provided from COXPRESdb. |
| Red dotted | PPI | PPI information is provided from TAIR and IntAct. |
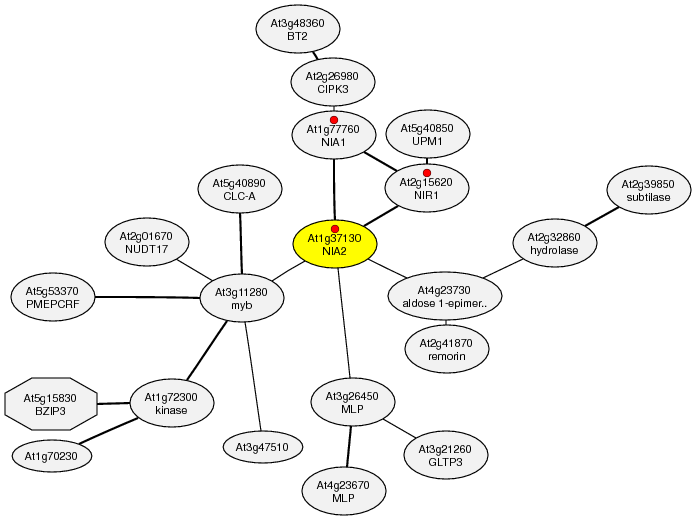
Example1; Coexpressed genes around At1g37130 (nitrate reductase 2 (NR2))
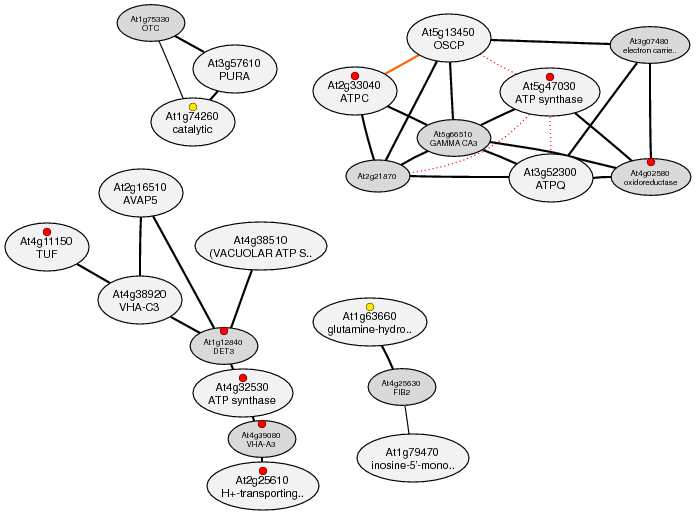
Example2; Coexpressed gene network for "Purineribonucleotide biosynthetic process" (GO:0009152)