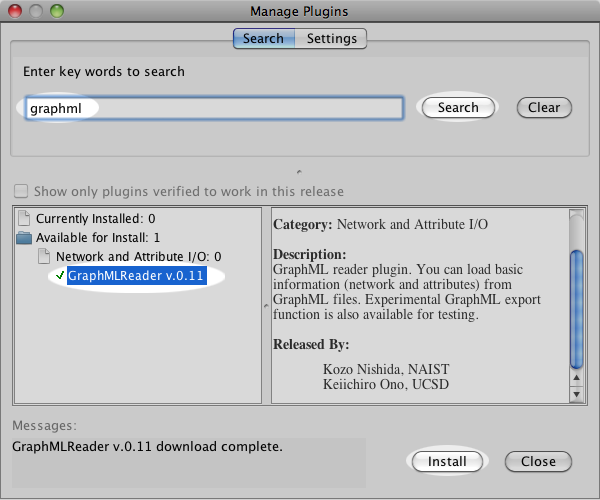
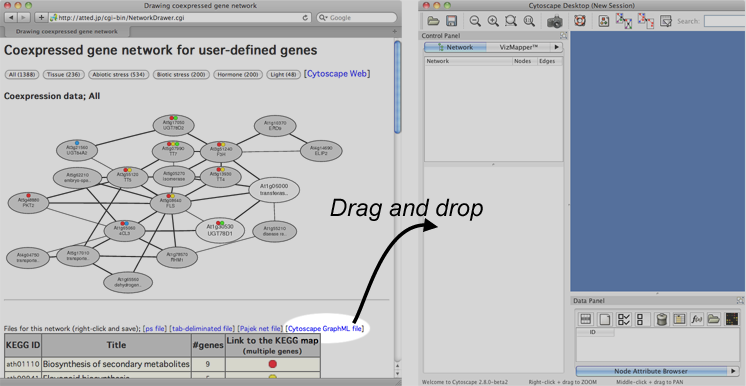
In the result page of NetworkDrawer, a link to GraphML file appears.
To open the network in Cytoscape, user can simply drag and drop the link of the GraphML file to Cytoscape.
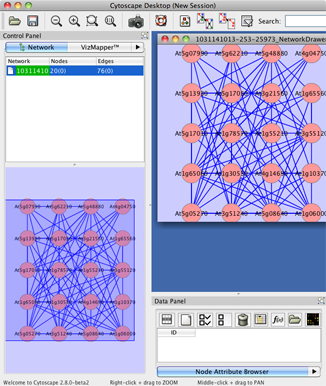
Then pre-layout network will appear. In the default, all of the edges appear, namely, coexpression in All, Tissue, Abiotic, Biotic, Hormone and Light conditions.
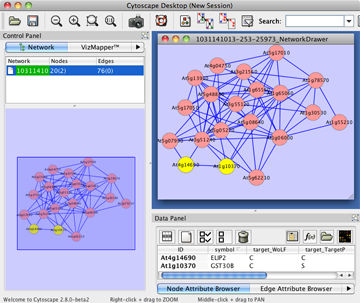
Cytoscape has many network layout functions. It can be used from Layout in the menu bar.
To know the detail functions of Cytoscape, please visit Cytoscape site (
www.cytoscape.org).