|
|
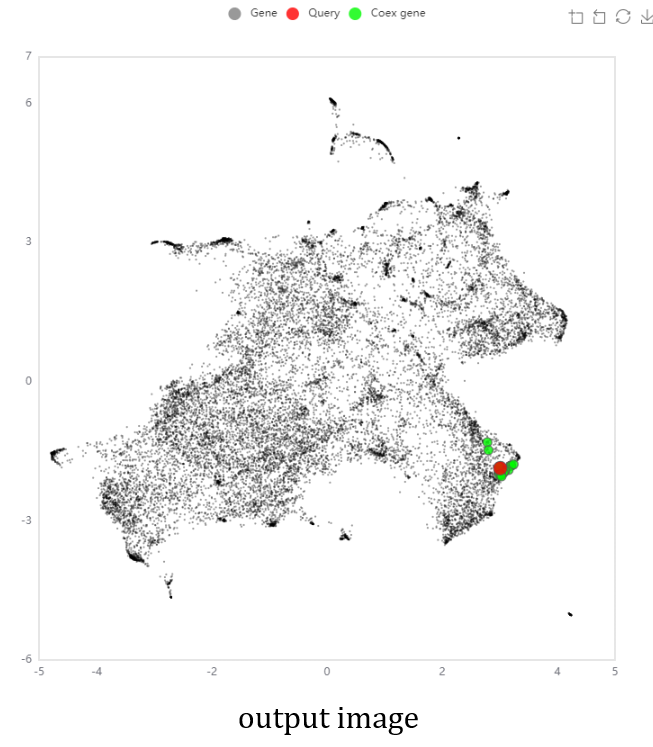
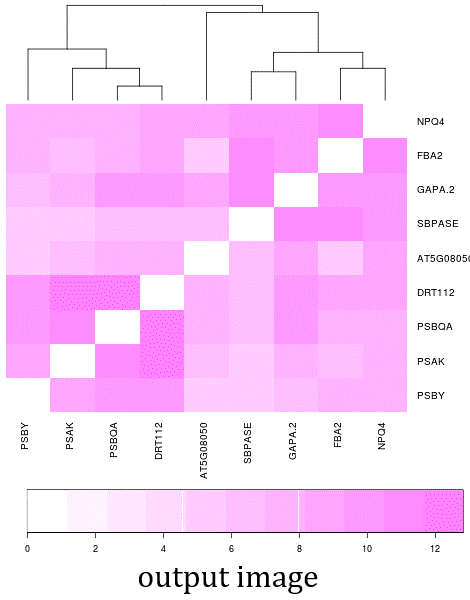
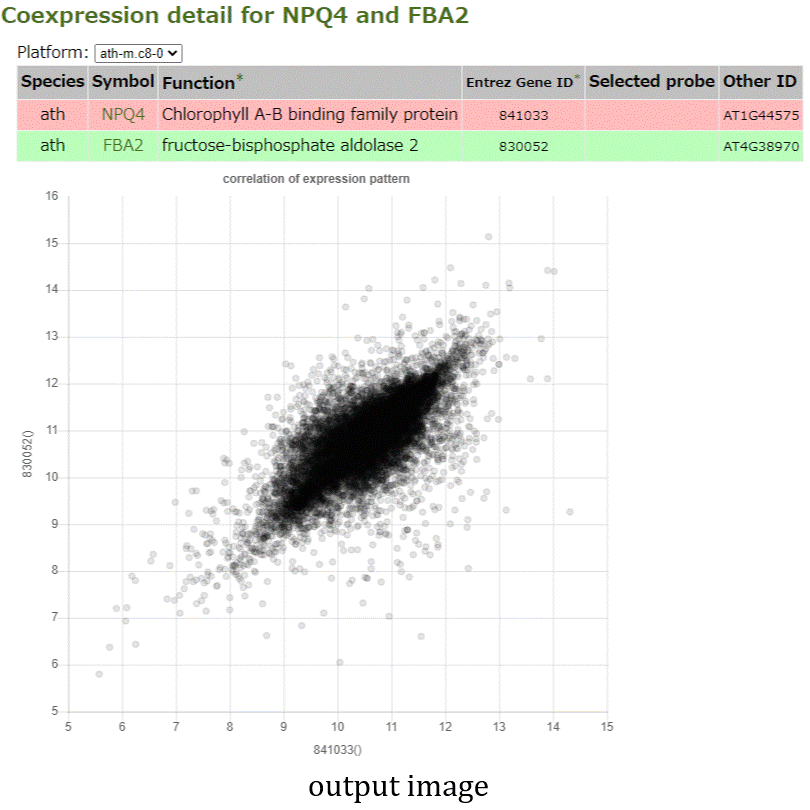
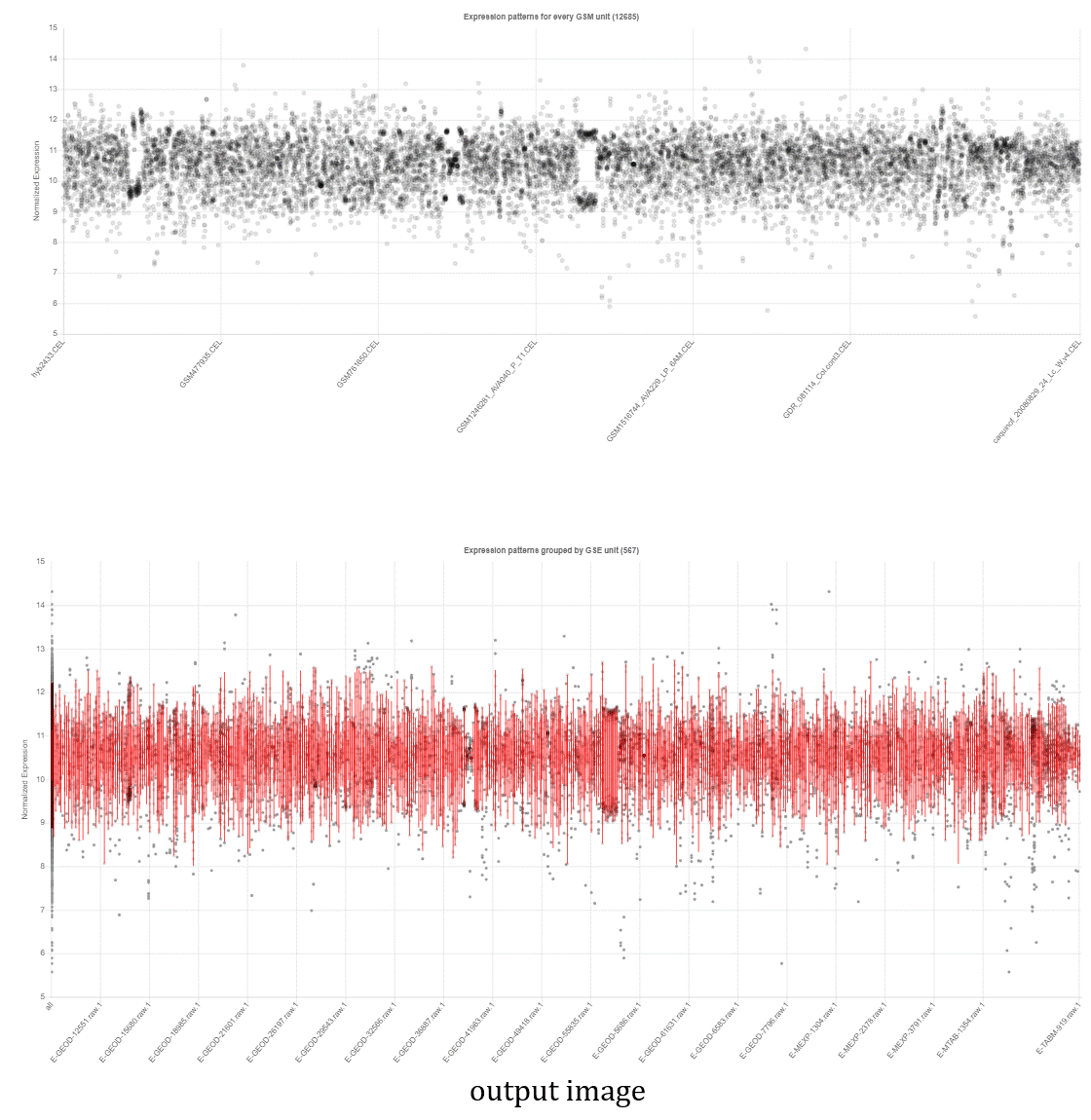
- The NetworkDrawer provides drawing and analyzing functions of coexpression networks for query genes.
- In addition to condition-independent coexpression, condition-specific coexpression data is drawn.
- INPUT: Delimitate loci with <RETURN> or <TAB>. Maximam number of input genes is limited to 100 for network drawing on ATTED-II. The example is genes annotated as "UDP-glycosyltransferase activity".
- OPTIONS:
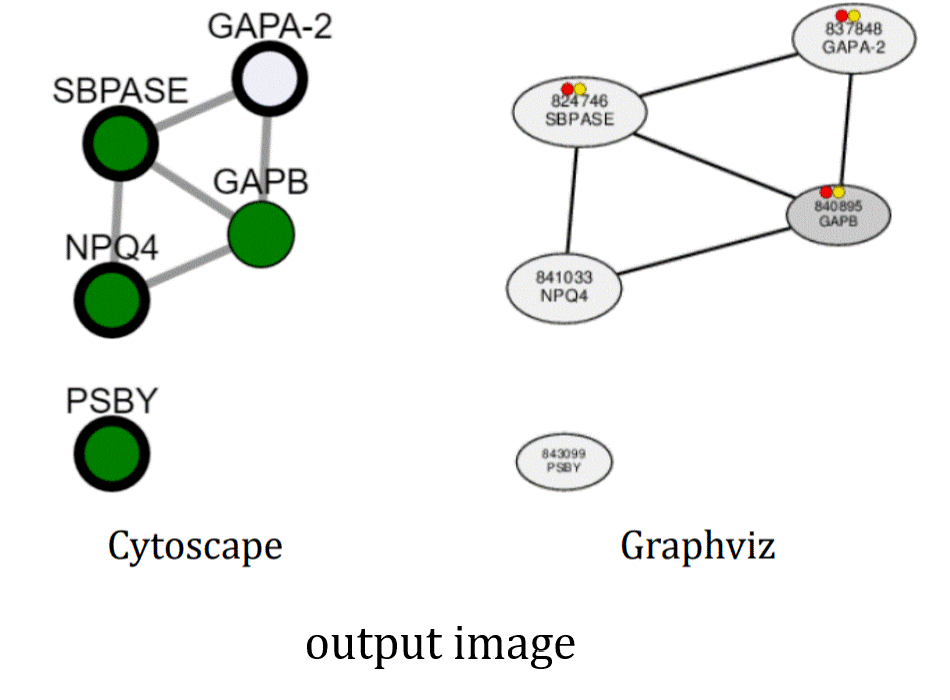
- Disp. type: When selecting "Graphviz", automatic subnetwork analyses are also provided to detect biologically-meaning subnetworks by applying enrichment tests for Gene Ontology annotations and heptamer cis elements in the procimal promoter region [-300, -1].
- Coex option: [Add no gene] drawing network only with the query genes. [Add a few genes] Automatically add genes connected with at least two query genes. [Add many genes] Automatically add genes around query genes. A group of the 20 coexpressed genes used in the coexpression network in corresponding locus page to the query genes are used. The nodes connected to a single node in the displaying
network are excluded for simplicity.
- PPIoption: [Do not draw PPIs] do not draw PPI in the gene network. [Draw PPIs] Drawer PPIs among the coexpressed genes. [Add a few genes] Automatically add genes connected with at least two genes in the network.
|