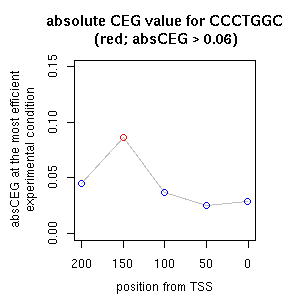
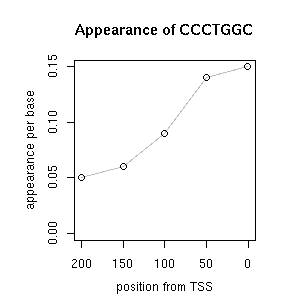
| reported seq | reported name | reference |
|---|---|---|
| no correspondence | ||
 |
 |
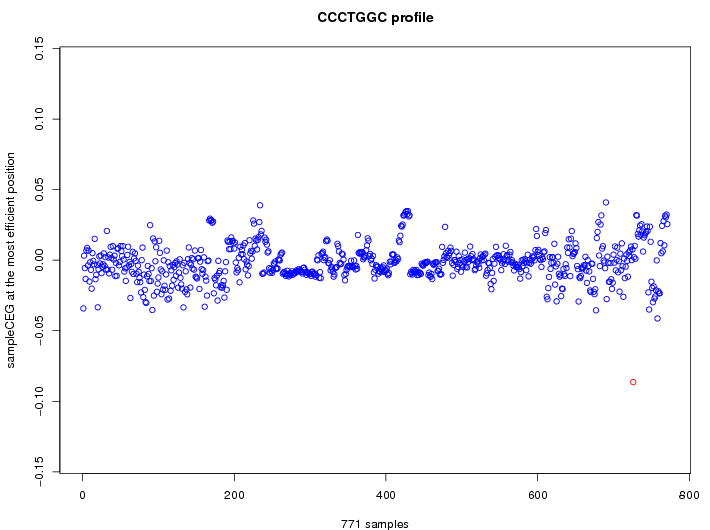
| GO ID | term | -Log(p) | number of {A; genes in a GO} | numver of {B; genes with this heptamer} | {A} x {B} |
|---|---|---|---|---|---|
| no functional bias detected | |||||
| correl | TF | function ((alias))* | TF family (link to AGRIS) |
|---|---|---|---|
| 0.46 | At4g16430 | basic helix-loop-helix (bHLH) family protein | BHLH |
| 0.46 | At5g06960 | bZIP family transcription factor (OBF5) ((OBF5, TGA5)) | bZIP |
| -0.46 | At3g58120 | bZIP transcription factor family protein | bZIP |
| -0.45 | At5g08330 | TCP family transcription factor, putative | TCP |
| -0.44 | At1g05805 | basic helix-loop-helix (bHLH) family protein | BHLH |
| 0.44 | At1g03040 | basic helix-loop-helix (bHLH) family protein | BHLH |
| 0.44 | At3g11200 | PHD finger family protein | Alfin-like |
| -0.43 | At1g68190 | zinc finger (B-box type) family protein | C2C2-CO-like |
| 0.42 | At5g18830 | squamosa promoter-binding protein-like 7 (SPL7) ((SPL7)) | SBP |
| 0.42 | At5g09330 | no apical meristem (NAM) family protein | NAC |
| 0.41 | At1g50420 | scarecrow-like transcription factor 3 (SCL3) ((SCL3)) | GRAS |
| 0.41 | At4g24470 | zinc finger (GATA type) protein ZIM (ZIM) ((ZIM)) | C2C2-Gata |
|