| functional annotation |
| Function |
myb domain protein 28 |




|
| GO BP |
|
GO:0010438 [list] [network] cellular response to sulfur starvation
|
(12 genes)
|
TAS
|
 |
|
GO:0010439 [list] [network] regulation of glucosinolate biosynthetic process
|
(13 genes)
|
IMP
|
 |
|
GO:0009625 [list] [network] response to insect
|
(33 genes)
|
IEP
|
 |
|
GO:0009682 [list] [network] induced systemic resistance
|
(34 genes)
|
IMP
|
 |
|
GO:0050832 [list] [network] defense response to fungus
|
(705 genes)
|
IMP
|
 |
|
GO:0009617 [list] [network] response to bacterium
|
(1192 genes)
|
IMP
|
 |
|
GO:0006355 [list] [network] regulation of DNA-templated transcription
|
(1656 genes)
|
IEP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0000976 [list] [network] transcription cis-regulatory region binding
|
(708 genes)
|
IPI
|
 |
|
GO:0003700 [list] [network] DNA-binding transcription factor activity
|
(1576 genes)
|
ISS
NAS
|
 |
|
| KEGG |
|
|
| Protein |
NP_200950.1
NP_851241.1
|
| BLAST |
NP_200950.1
NP_851241.1
|
| Orthologous |
[Ortholog page]
LOC103860592 (bra)
|
Subcellular
localization
wolf |
|
nucl 7,
cyto 1,
extr 1,
pero 1,
cyto_E.R. 1,
cyto_plas 1
|
(predict for NP_200950.1)
|
|
nucl 8,
cyto_nucl 4,
mito 1,
plas 1,
mito_plas 1
|
(predict for NP_851241.1)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_200950.1)
|
|
mito 7,
other 3
|
(predict for NP_851241.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00966 |
Glucosinolate biosynthesis |
8 |
 |
| ath01210 |
2-Oxocarboxylic acid metabolism |
8 |
 |
| ath00290 |
Valine, leucine and isoleucine biosynthesis |
3 |
 |
Genes directly connected with MYB28 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 12.1 |
CYP83A1 |
cytochrome P450, family 83, subfamily A, polypeptide 1 |
[detail] |
827011 |
| 11.6 |
BAT5 |
bile acid transporter 5 |
[detail] |
826811 |
| 11.0 |
IPMI1 |
isopropylmalate isomerase 1 |
[detail] |
825068 |
| 6.4 |
PTR4 |
Major facilitator superfamily protein |
[detail] |
814733 |
| 5.8 |
HMA2 |
heavy metal atpase 2 |
[detail] |
829134 |
| 5.5 |
AT5G03995 |
uncharacterized protein |
[detail] |
6240402 |
| 5.2 |
AT3G45210 |
transcription initiation factor TFIID subunit (Protein of unknown function, DUF584) |
[detail] |
823657 |
| 5.1 |
ACA1 |
alpha carbonic anhydrase 1 |
[detail] |
824438 |
|
Coexpressed
gene list |
[Coexpressed gene list for MYB28]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
247549_at
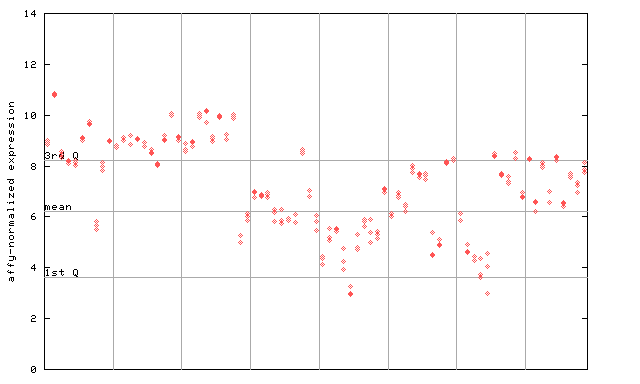
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
247549_at
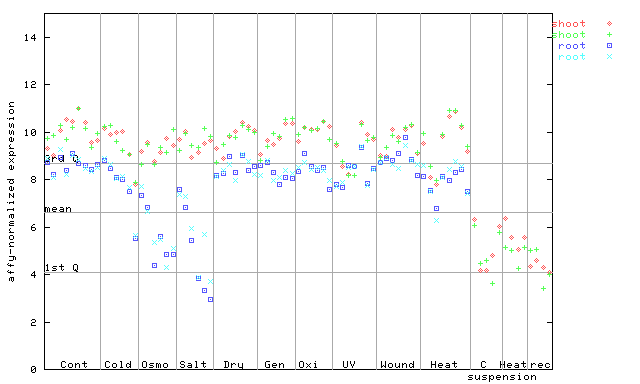
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
247549_at
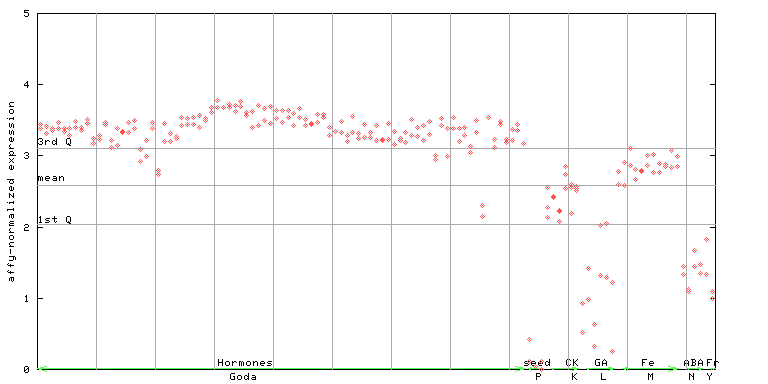
X axis is samples (xls file), and Y axis is log-expression.
|















