[←][→] ath
| functional annotation | ||||||||
| Function | zinc finger CCCH domain protein |




|
||||||
| GO BP |
|
|||||||
| GO CC |
|
|||||||
| GO MF |
|
|||||||
| KEGG | ||||||||
| Protein | NP_001322669.1 NP_564158.1 NP_973881.2 | |||||||
| BLAST | NP_001322669.1 NP_564158.1 NP_973881.2 | |||||||
| Orthologous | [Ortholog page] LOC9272415 (osa) LOC11442269 (mtr) LOC18099517 (ppo) LOC18109702 (ppo) LOC100306341 (gma) LOC100778362 (gma) LOC101251864 (sly) LOC103869075 (bra) LOC123050118 (tae) LOC123125770 (tae) LOC123181455 (tae) LOC123436008 (hvu) LOC123436024 (hvu) | |||||||
| Subcellular localization wolf |
|
|||||||
| Subcellular localization TargetP |
|
|||||||
| Gene coexpression | ||||||||
| Network*for coexpressed genes |
||||||||
|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for AT1G22140] | |||||||
| Gene expression | ||||||||
| All samples | [Expression pattern for all samples] | |||||||
| AtGenExpress* (Development) |
255960_at
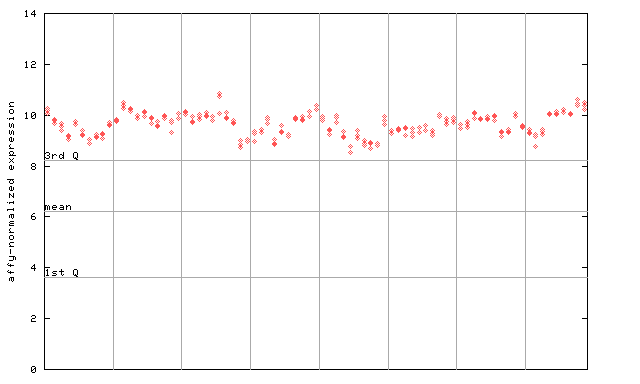
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||
| AtGenExpress* (Stress) |
255960_at
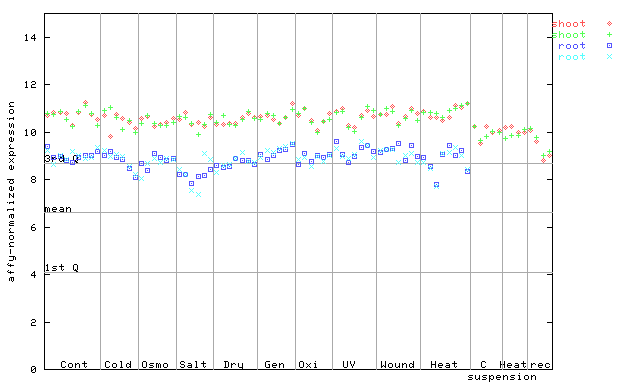
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||
| AtGenExpress* (Hormone) |
255960_at
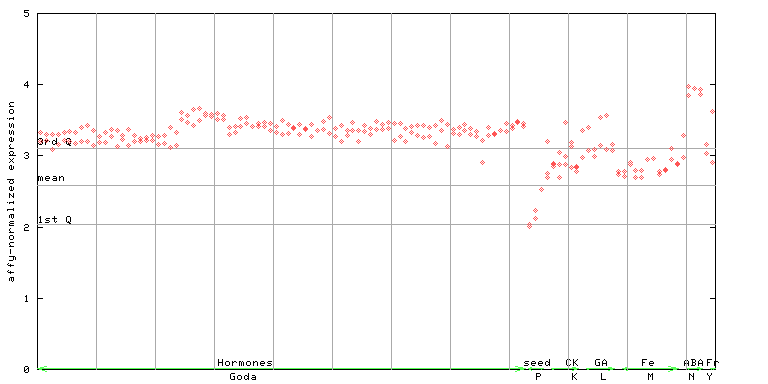
X axis is samples (xls file), and Y axis is log-expression. |
|||||||
| Link to other DBs | ||
| Entrez Gene ID | 838819 |


|
| Refseq ID (protein) | NP_001322669.1 |  |
| NP_564158.1 |  |
|
| NP_973881.2 |  |
|
The preparation time of this page was 0.1 [sec].

