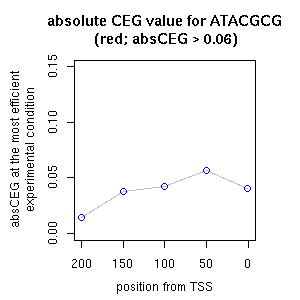
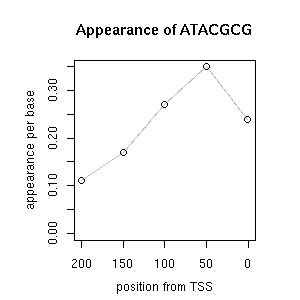
| reported seq | reported name | reference |
|---|---|---|
| no correspondence | ||
 |
 |

| GO ID | term | -Log(p) | number of {A; genes in a GO} | numver of {B; genes with this heptamer} | {A} x {B} |
|---|---|---|---|---|---|
| no functional bias detected | |||||
| correl | TF | function ((alias))* | TF family (link to AGRIS) |
|---|---|---|---|
| 0.64 | At5g01380 | expressed protein | Trihelix |
| 0.62 | At5g24590 | turnip crinkle virus-interacting protein / TCV-interacting protein (TIP) ((TIP)) | NAC |
| 0.61 | At2g38250 | DNA-binding protein-related | Trihelix |
| 0.59 | At4g01250 | WRKY family transcription factor ((WRKY22)) | WRKY |
| 0.58 | At5g48655 | zinc finger (C3HC4-type RING finger) family protein | C3H |
| 0.57 | At1g22810 | AP2 domain-containing transcription factor, putative | AP2-EREBP |
| 0.56 | At5g49520 | WRKY family transcription factor ((WRKY48)) | WRKY |
| 0.55 | At1g28370 | ERF domain protein 11 (ERF11) | AP2-EREBP |
| 0.54 | At5g45710 | heat shock transcription factor family protein ((AT-HSFA4C)) | HSF |
| 0.53 | At3g62420 | bZIP transcription factor family protein | bZIP |
| 0.53 | At3g15210 | ethylene-responsive element-binding factor 4 (ERF4) ((ATERF4, RAP2.5)) | AP2-EREBP |
| 0.52 | At3g23250 | myb family transcription factor (MYB15) | MYB |
| 0.52 | At3g10800 | bZIP transcription factor family protein | bZIP |
| 0.52 | At4g33940 | zinc finger (C3HC4-type RING finger) family protein | C3H |
| 0.51 | At1g80840 | WRKY family transcription factor ((WRKY40)) | WRKY |
|