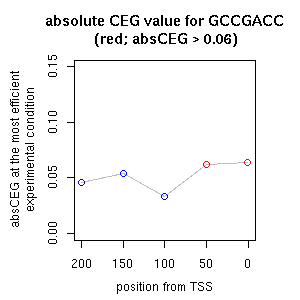
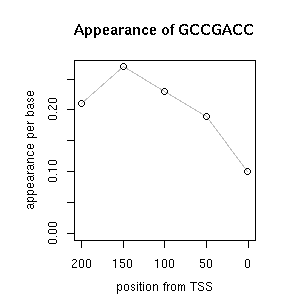
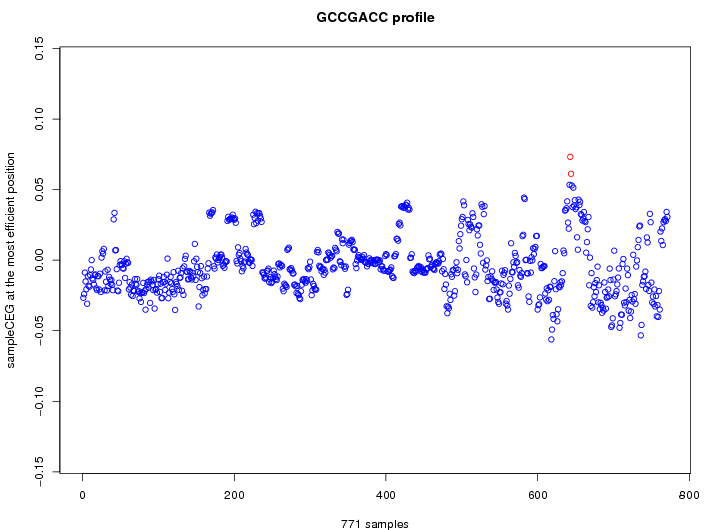
| reported seq | reported name | reference |
|---|---|---|
| RCCGAC | DRECRTCOREAT | [ref] [ref] [ref] [ref] [ref] |
| CCGAC | LTRECOREATCOR15 | [ref] [ref] [ref] [ref] |
 |
 |
| GO ID | term | -Log(p) | number of {A; genes in a GO} | numver of {B; genes with this heptamer} | {A} x {B} |
|---|---|---|---|---|---|
| no functional bias detected | |||||
| correl | TF | function ((alias))* | TF family (link to AGRIS) |
|---|---|---|---|
| 0.52 | At2g46830 | myb-related transcription factor (CCA1) ((CCA1)) | MYB-related |
| 0.49 | At5g57660 | zinc finger (B-box type) family protein | C2C2-CO-like |
| 0.48 | At3g47500 | Dof-type zinc finger domain-containing protein | C2C2-Dof |
| 0.42 | At5g15850 | zinc finger protein CONSTANS-LIKE 1 (COL1) ((COL1)) | C2C2-CO-like |
| 0.41 | At5g62430 | Dof-type zinc finger domain-containing protein | C2C2-Dof |
| 0.40 | At5g60850 | Dof-type zinc finger domain-containing protein ((OBP4)) | C2C2-Dof |
|