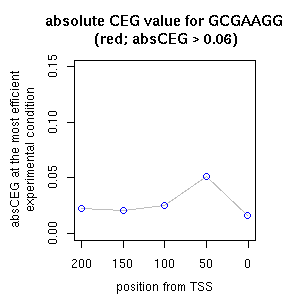
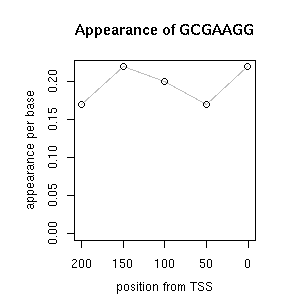
| correl | TF | function ((alias))* | TF family
(link to AGRIS) |
|---|
| -0.48 | At3g19580 | zinc finger (C2H2 type) protein 2 (AZF2) ((AZF2))
| C2H2
|
| -0.46 | At3g52800 | zinc finger (AN1-like) family protein
| C2H2
|
| -0.45 | At5g13820 | telomeric DNA-binding protein 1 (TBP1) ((HPPBF-1, TBP1))
| MYB-related
|
| -0.44 | At4g37790 | homeobox-leucine zipper protein 22 (HAT22) / HD-ZIP protein 22 ((HAT22))
| Homeobox
|
| -0.44 | At1g62300 | WRKY family transcription factor ((WRKY6))
| WRKY
|
| -0.44 | At3g16720 | zinc finger (C3HC4-type RING finger) family protein ((ATL2))
| C3H
|
| -0.43 | At2g38470 | WRKY family transcription factor ((WRKY33))
| WRKY
|
| -0.43 | At1g69490 | no apical meristem (NAM) family protein ((NAP))
| NAC
|
| -0.43 | At1g42990 | bZIP transcription factor family protein
| bZIP
|
| -0.43 | At3g15210 | ethylene-responsive element-binding factor 4 (ERF4) ((ATERF4, RAP2.5))
| AP2-EREBP
|
| -0.43 | At1g77450 | no apical meristem (NAM) family protein
| NAC
|
| -0.42 | At3g04060 | no apical meristem (NAM) family protein
| NAC
|
| -0.42 | At3g49530 | no apical meristem (NAM) family protein
| NAC
|
| -0.42 | At3g04070 | no apical meristem (NAM) family protein
| NAC
|
| -0.42 | At5g63790 | no apical meristem (NAM) family protein
| NAC
|