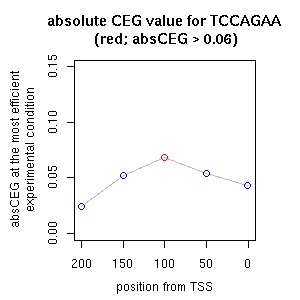
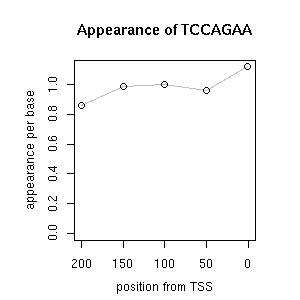
| correl | TF | function ((alias))* | TF family
(link to AGRIS) |
|---|
| 0.64 | At2g26150 | heat shock transcription factor family protein ((ATHSFA2))
| HSF
|
| 0.63 | At5g04410 | no apical meristem (NAM) family protein ((NAC2))
| NAC
|
| 0.58 | At4g11660 | heat shock factor protein 7 (HSF7) / heat shock transcription factor 7 (HSTF7) ((AT-HSFB2B))
| HSF
|
| 0.58 | At1g26800 | zinc finger (C3HC4-type RING finger) family protein
| C3H
|
| 0.58 | At5g05410 | DRE-binding protein (DREB2A) ((DREB2A))
| AP2-EREBP
|
| 0.57 | At1g14200 | zinc finger (C3HC4-type RING finger) family protein
| C3H
|
| 0.53 | At4g36990 | heat shock factor protein 4 (HSF4) / heat shock transcription factor 4 (HSTF4) ((AT-HSFB1, ATHSF4, HSF4))
| HSF
|
| 0.49 | At3g10800 | bZIP transcription factor family protein
| bZIP
|
| 0.48 | At5g62020 | heat shock factor protein, putative (HSF6) / heat shock transcription factor, putative (HTSF6) ((AT-HSFB2A))
| HSF
|
| 0.47 | At1g32870 | no apical meristem (NAM) family protein
| NAC
|
| 0.47 | At3g63350 | heat shock transcription factor family protein ((AT-HSFA7B))
| HSF
|
| 0.46 | At4g00940 | Dof-type zinc finger domain-containing protein
| C2C2-Dof
|
| 0.45 | At2g40950 | bZIP transcription factor family protein
| bZIP
|
| 0.45 | At1g01720 | no apical meristem (NAM) family protein ((ATAF1))
| NAC
|
| 0.45 | At1g56170 | transcription factor, putative ((HAP5B))
| CCAAT-HAP5
|