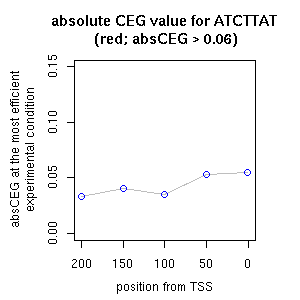
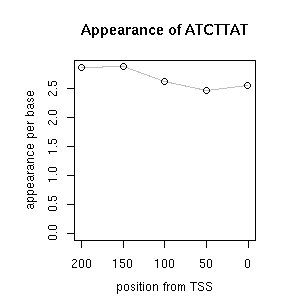
| reported seq | reported name | reference |
|---|---|---|
| ATCTTATGTCATTGATGACGACCTCC | OBF5ATGST6 | [ref] [ref] [ref] [ref] |
 |
 |
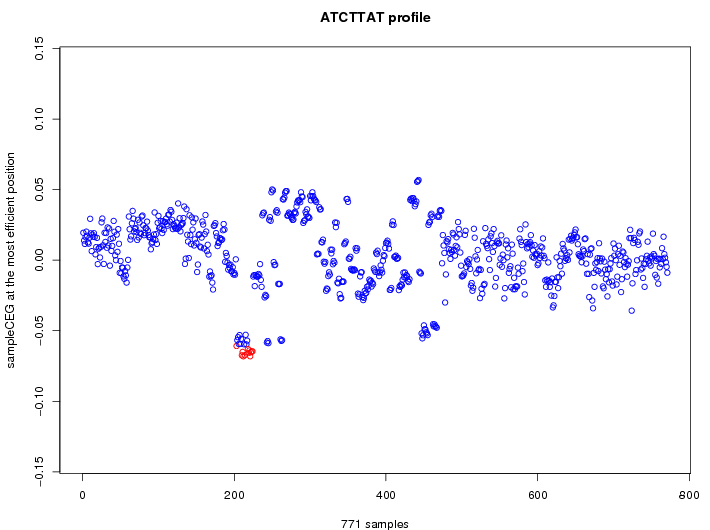
| GO ID | term | -Log(p) | number of {A; genes in a GO} | numver of {B; genes with this heptamer} | {A} x {B} |
|---|---|---|---|---|---|
| no functional bias detected | |||||
| correl | TF | function ((alias))* | TF family (link to AGRIS) |
|---|---|---|---|
| 0.64 | At4g14540 | CCAAT-box binding transcription factor subunit B (NF-YB) (HAP3 ) (AHAP3) family | CCAAT-HAP3 |
| 0.63 | At1g68520 | zinc finger (B-box type) family protein | C2C2-CO-like |
| 0.58 | At1g68190 | zinc finger (B-box type) family protein | C2C2-CO-like |
| 0.54 | At2g18300 | basic helix-loop-helix (bHLH) family protein | BHLH |
| 0.52 | At5g56860 | zinc finger (GATA type) family protein | C2C2-Gata |
| 0.51 | At5g60970 | TCP family transcription factor, putative | TCP |
| 0.50 | At2g02450 | no apical meristem (NAM) family protein | NAC |
| 0.49 | At1g34310 | transcriptional factor B3 family protein / auxin-responsive factor AUX/IAA-related | ARF |
| 0.49 | At4g36540 | basic helix-loop-helix (bHLH) family protein | BHLH |
| 0.49 | At2g20180 | basic helix-loop-helix (bHLH) family protein ((PIL5)) | BHLH |
| 0.48 | At3g59060 | basic helix-loop-helix (bHLH) family protein ((PIL6)) | BHLH |
| 0.48 | At2g41940 | zinc finger (C2H2 type) family protein ((ZFP8)) | C2H2 |
| 0.48 | At2g20570 | golden2-like transcription factor (GLK1) ((GLK1, GPRI1)) | G2-like |
| 0.47 | At4g01460 | basic helix-loop-helix (bHLH) family protein | BHLH |
| 0.46 | At5g20220 | zinc knuckle (CCHC-type) family protein | C2H2 |
|