| reported seq | reported name | reference |
|---|---|---|
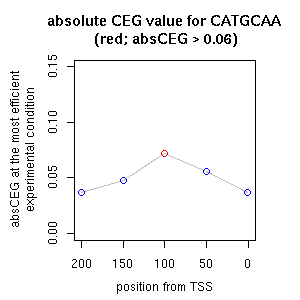
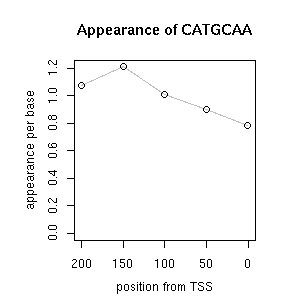
| TCCATAGCCATGCAWRCTGMAGAATGTC | LEGUMINBOXLEGA5 | [ref] [ref] |
| CATGCA | RYREPEATBNNAPA | [ref] [ref] |
 |
 |
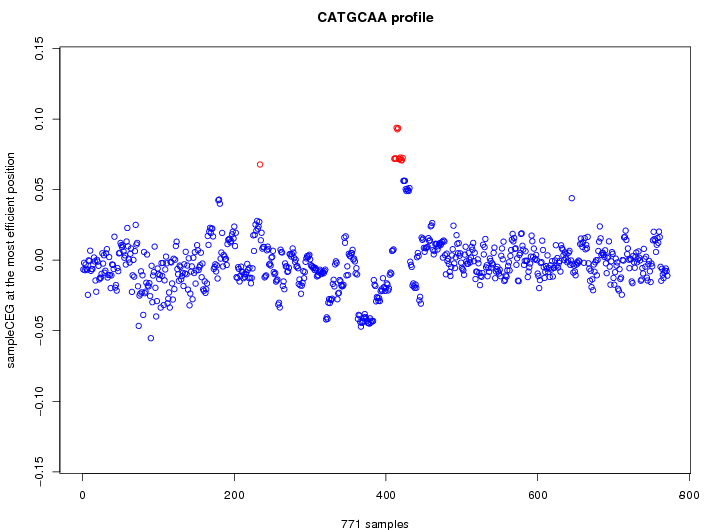
| GO ID | term | -Log(p) | number of {A; genes in a GO} | numver of {B; genes with this heptamer} | {A} x {B} |
|---|---|---|---|---|---|
| no functional bias detected | |||||
| correl | TF | function ((alias))* | TF family (link to AGRIS) |
|---|---|---|---|
| 0.53 | At3g24650 | abscisic acid-insensitive protein 3 (ABI3) ((ABI3)) | ABI3VP1 |
| 0.51 | At5g07500 | zinc finger (CCCH-type) family protein ((PEI1)) | C2H2 |
| 0.50 | At3g26790 | transcriptional regulator (FUSCA3) ((FUS3)) | ABI3VP1 |
| -0.50 | At5g64340 | expressed protein | BHLH |
| -0.48 | At4g30930 | 50S ribosomal protein L21, mitochondrial (RPL21M) ((WRKY32)) | WRKY |
| -0.48 | At5g09460 | expressed protein | BHLH |
| 0.48 | At3g56660 | bZIP transcription factor family protein | bZIP |
| -0.47 | At5g16070 | chaperonin, putative | TCP |
| 0.46 | At1g75490 | DRE-binding transcription factor, putative | AP2-EREBP |
| 0.45 | At5g47670 | CCAAT-box binding transcription factor family protein / leafy cotyledon 1-related (L1L) | CCAAT-HAP3 |
| -0.45 | At4g13640 | myb family transcription factor | G2-like |
| 0.45 | At3g15510 | no apical meristem (NAM) family protein (NAC2) ((ATNAC2)) | NAC |
| 0.45 | At1g77950 | MADS-box family protein | MADS |
| 0.45 | At3g44460 | basic leucine zipper transcription factor (BZIP67) ((DPBF2)) | bZIP |
| 0.45 | At3g13540 | myb family transcription factor ((ATMYB5)) | MYB |
|