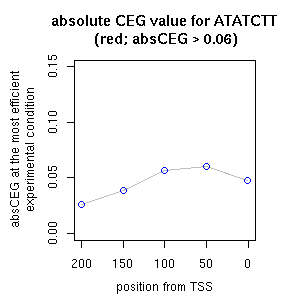
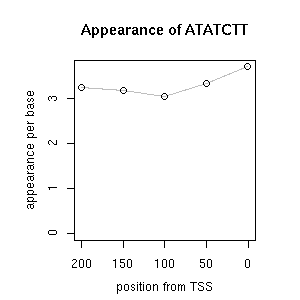
| reported seq | reported name | reference |
|---|---|---|
| no correspondence | ||
 |
 |
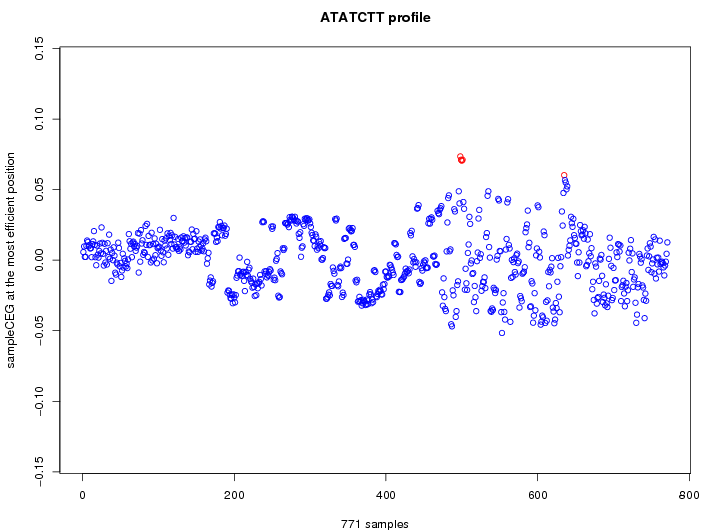
| GO ID | term | -Log(p) | number of {A; genes in a GO} | numver of {B; genes with this heptamer} | {A} x {B} |
|---|---|---|---|---|---|
| no functional bias detected | |||||
| correl | TF | function ((alias))* | TF family (link to AGRIS) |
|---|---|---|---|
| 0.59 | At1g22070 | bZIP family transcription factor (TGA3) ((TGA3)) | bZIP |
| 0.59 | At1g67970 | heat shock factor protein, putative (HSF5) / heat shock transcription factor, putative (HSTF5) ((AT-HSFA8)) | HSF |
| -0.58 | At1g01060 | myb family transcription factor ((LHY)) | MYB-related |
| 0.58 | At1g49720 | ABA-responsive element-binding protein / abscisic acid responsive elements-binding factor (ABRE) ((ABF1)) | bZIP |
| 0.58 | At1g05805 | basic helix-loop-helix (bHLH) family protein | BHLH |
| 0.57 | At1g78600 | zinc finger (B-box type) family protein | C2C2-CO-like |
| 0.57 | At3g50700 | zinc finger (C2H2 type) family protein | C2H2 |
| 0.56 | At2g18670 | zinc finger (C3HC4-type RING finger) family protein | C3H |
| 0.55 | At5g05090 | myb family transcription factor | G2-like |
| -0.55 | At4g38960 | zinc finger (B-box type) family protein | C2C2-CO-like |
| 0.54 | At2g47890 | zinc finger (B-box type) family protein | C2C2-CO-like |
| 0.53 | At1g08900 | sugar transporter-related | C2H2 |
| 0.48 | At3g45260 | zinc finger (C2H2 type) family protein | C2H2 |
| -0.48 | At5g53200 | myb family transcription factor (TRIPTYCHON) | MYB |
| 0.48 | At5g48250 | zinc finger (B-box type) family protein | C2C2-CO-like |
|